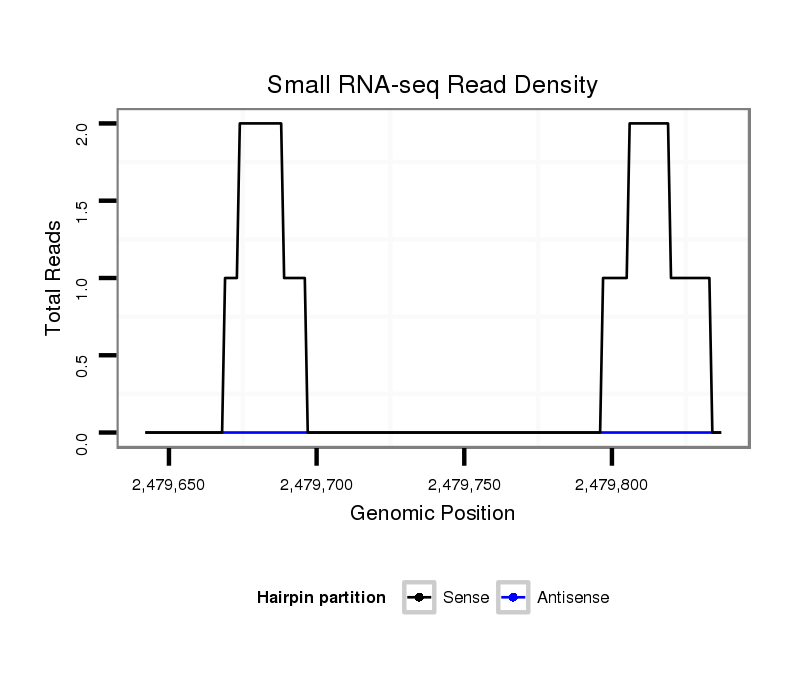
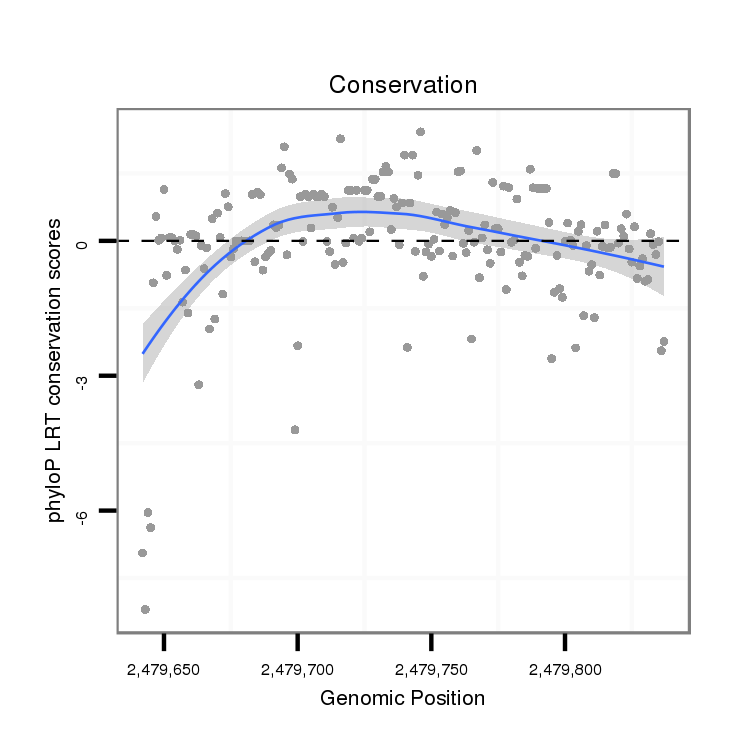
| CTGC---------------------TG-------------ATGCTTCCGCCACTAC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTGTTGTTGC---------TGCTGGA---------------------GTTGTT------------GCTGATGCTGCTG------------AGGCAGCCGCCGCT---GTCGCC-----------GCTGGTGCCCCCGCTGCCGTC-------CCGGAGCGAT-----------------------------------------------CTGCCGT------------------------------------CTCGATGCGCAGTC-TTGA----------------------------------CTGTG-------------------------------------------------------------------------------------------TCA-TCAAG--AAT---GGAGCTGACGGCGTTATGGAGGAAGCGGCCA---------------------------------GCATCAAG----G | Size | Hit Count | Total Norm | Total | M044
Female-body | M058
Embryo | V039
Embryo | V055
Head | V105
Male-body | V106
Head |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................GCCGT------------------------------------CTCGATGCGCAGTC-TTGA----------------------------------.............................................................................................................................................................................................. | 23 | 15 | 1.07 | 16 | 13 | 1 | 2 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................GTT------------GCTGATGCTGCTG------------AGGCAGCC............................................................................................................................................................................................................................................................................................................................................................................................................. | 24 | 20 | 0.95 | 19 | 18 | 1 | 0 | 0 | 0 | 0 |
| ....................................................CTAC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTGT........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.95 | 19 | 17 | 2 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................GCCGT------------------------------------CTCGATGCGCAGTC-TTG................................................................................................................................................................................................................................. | 22 | 15 | 0.93 | 14 | 13 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................GTT------------GCTGATGCTGCTG------------AGGCAGCCG............................................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 20 | 0.90 | 18 | 18 | 0 | 0 | 0 | 0 | 0 |
| ....................................................CTAC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTGTTG......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.85 | 17 | 17 | 0 | 0 | 0 | 0 | 0 |
| ....................................................CTAC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTG............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 20 | 0.65 | 13 | 13 | 0 | 0 | 0 | 0 | 0 |
| ....................................................CTAC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTGTT.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.50 | 10 | 10 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................GTT------------GCTGATGCTGCTG------------AGGCAG............................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.45 | 9 | 9 | 0 | 0 | 0 | 0 | 0 |
| ....................................................CTAC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTGTTGTTGC---------............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 28 | 20 | 0.40 | 8 | 8 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAAG--AAT---GGAGCTGACGGCGTTA............................................................. | 23 | 13 | 0.31 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ....................................................CTAC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTGTTGTTG...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................GCCGT------------------------------------CTCGATGCGCAGTC-TTGA----------------------------------C............................................................................................................................................................................................. | 24 | 15 | 0.27 | 4 | 3 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................GCCGT------------------------------------CTCGATGCGCAGTC-TT.................................................................................................................................................................................................................................. | 21 | 15 | 0.27 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| .....................................................TAC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTGTTG......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAAG--AAT---GGAGCTGACGGCGTTAT............................................................ | 24 | 13 | 0.23 | 3 | 1 | 2 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCA-TCAAG--AAT---GGAGCTGACGG.................................................................. | 22 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAAG--AAT---GGAGCTGACGGCGT............................................................... | 21 | 20 | 0.20 | 4 | 3 | 0 | 1 | 0 | 0 | 0 |
| ....................................................CTAC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTGTTGT........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................CCGT------------------------------------CTCGATGCGCAGTC-TTGA----------------------------------.............................................................................................................................................................................................. | 22 | 15 | 0.20 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAAG--AAT---GGAGCTGACGGCGTTATGGA......................................................... | 27 | 13 | 0.15 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCAAG--AAT---GGAGCTGACGGCGTTA............................................................. | 24 | 13 | 0.15 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAAG--AAT---GGAGCTGACGGCGTTATG........................................................... | 25 | 13 | 0.15 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCAAG--AAT---GGAGCTGACGGCGTTT............................................................. | 24 | 13 | 0.15 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................TAC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTGT........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................TT------------GCTGATGCTGCTG------------AGGCAGCCGC........................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................GCCGT------------------------------------CTCGATGCGCAGTC-TTGA----------------------------------CTG........................................................................................................................................................................................... | 26 | 15 | 0.13 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................CGT------------------------------------CTCGATGCGCAGTC-TTGA----------------------------------.............................................................................................................................................................................................. | 21 | 15 | 0.13 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................CCGT------------------------------------CTCGATGCGCAGTC-TTG................................................................................................................................................................................................................................. | 21 | 16 | 0.13 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .....................................................TAC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTGTTGTTG...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCA-TCAAG--AAT---GGAGCTGACGGC................................................................. | 23 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................GCT---GTCGCC-----------GCTGGTGCCCCCGCTGC.................................................................................................................................................................................................................................................................................................................................................................. | 26 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAAG--AAT---GGAGCTGACGG.................................................................. | 18 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................GCCGT------------------------------------CTCGATGCGCAGTC-TTGA----------------------------------CTGTG-------------------------------------------------------------------------------------------.............................................................................................. | 28 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................GT------------------------------------CTCGATGCGCAGTC-TTGA----------------------------------CTGTG-------------------------------------------------------------------------------------------TC............................................................................................ | 27 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CTGTG-------------------------------------------------------------------------------------------TCA-TCAAG--AAT---GGAGC........................................................................ | 21 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TG-------------------------------------------------------------------------------------------TCA-TCAAG--AAT---GGAGCTGACGG.................................................................. | 24 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................G-------------------------------------------------------------------------------------------TCA-TCAAG--AAT---GGAGCTGACGGCG................................................................ | 25 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AAG--AAT---GGAGCTGACGGCGTTAT............................................................ | 23 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................T---GGAGCTGACGGCGTTATGGAGGAAGC................................................... | 27 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAAG--AAT---GGAGCTGACGGCGTT.............................................................. | 22 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AAG--AAT---GGAGCTGACGGCGTTATGG.......................................................... | 25 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCAAG--AAT---GGAGCTGACGGCGTTATGG.......................................................... | 27 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CTGACGGCGTTATGGAGGAAGCGGCC............................................... | 26 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................A-TCAAG--AAT---GGAGCTGACGGCGTTATG........................................................... | 27 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCAAG--AAT---GGAGCTGACGGCGTTATG........................................................... | 26 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................GCCGT------------------------------------CTCGATGCGCAGTC-TTC................................................................................................................................................................................................................................. | 22 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................T-----------------------------------------------------------G------------------------CCCC------CGCTGTTGTTGC---------TGCTGGA---------------------................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................TGCCGT------------------------------------CTCGATGCGCAGTC-TTGA----------------------------------.............................................................................................................................................................................................. | 24 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................T-----------------------------------------------------------G------------------------CCCC------CGCTGTTGTTGC---------TGC......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................CCGT------------------------------------CTCGATGCGCAGTC-TT.................................................................................................................................................................................................................................. | 20 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................GC---------TGCTGGA---------------------GTTGTT------------GCTGATG....................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................AC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTGT........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................GAT-----------------------------------------------CTGCCGT------------------------------------CTCGATGCGC......................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................TGGTGCCCCCGCTGCCGTC-------CCGGAGC................................................................................................................................................................................................................................................................................................................................................ | 26 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCAAG--AAT---GGAGCTGACGGCGT............................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................AC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTGTTGTTG...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................T---GTCGCC-----------GCTGGTGCCCCCGCTGCCGT............................................................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................TT------------GCTGATGCTGCTG------------AGGCAGCC............................................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................GCT---GTCGCC-----------GCTGGTGCCCCCGCTGCCG................................................................................................................................................................................................................................................................................................................................................................ | 28 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................AC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTGTTG......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................TCGCC-----------GCTGGTGCCCCCGCTGC.................................................................................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................TGCTGGA---------------------GTTGTT------------GCTGATGCTGCA.................................................................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CA-TCAAG--AAT---GGAGCTGACGGC................................................................. | 22 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................TC-------CCGGAGCGAT-----------------------------------------------CTGCCGT------------------------------------CTCGATG............................................................................................................................................................................................................................................ | 26 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................TT------------GCTGATGCTGCTG------------AGGCA................................................................................................................................................................................................................................................................................................................................................................................................................ | 20 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................CTGCCGTC-------CCGGAGCGAT-----------------------------------------------CTGCCGT------------------------------------................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................................................................................................................................................................................................................CT---GTCGCC-----------GCTGGTGCCCCCGCTGC.................................................................................................................................................................................................................................................................................................................................................................. | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................TGCT-----------------------------------------------------------G------------------------CCCC------CGCTGTTGTTGC---------TG.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCAAG--AAT---GGAGCTGACGGCG................................................................ | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................AC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTGTTGT........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 23 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................CTGGA---------------------GTTGTT------------GCTGATGCTGCT.................................................................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................TGCTGGA---------------------GTTGTT------------GCTGATGCTG.................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................GTT------------GCTGATGCTGCTG------------AGGCAGCCGT........................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................TAC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTGTTGT........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................CGGAGCGAT-----------------------------------------------CTGCCGT------------------------------------CTCGATG............................................................................................................................................................................................................................................ | 23 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCA-TCAAG--AAT---GGAGCTGACGGCGC............................................................... | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................TAC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCT............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................C-------CCGGAGCGAT-----------------------------------------------CTGCCGT------------------------------------CTC................................................................................................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................CTAC------------------------------------CTTTGCT-----------------------------------------------------------G------------------------CCCC------CGCTGTTGTT....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCAAG--AAT---GGAGCTGACGG.................................................................. | 19 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................T---GTCGCC-----------GCTGGTGCCCCCGCTGCC................................................................................................................................................................................................................................................................................................................................................................. | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................GCCGCCGCT---GTCGCC-----------GCTGGTGCCC......................................................................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................GTT------------GCTGATGCTGCTG------------AGGCAGCCGCC.......................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................TGCTGGA---------------------GTTGTT------------GCTGATGCTGCTG------------..................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................GTT------------GCTGATGCTGCTG------------AGGCA................................................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................TGGTGCCCCCGCTGCCGTC-------CCGG................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................TGTT------------GCTGATGCTGCTG------------AGGCAGTT............................................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................GTC-------CCGGAGCGAT-----------------------------------------------CTGCCGT------------------------------------CTCGAT............................................................................................................................................................................................................................................. | 26 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |