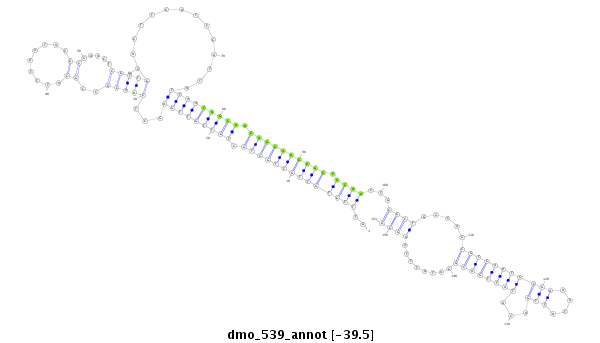

ID:dmo_539 |
Coordinate:scaffold_6359:417694-417844 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -39.4 | -39.2 | -39.1 |
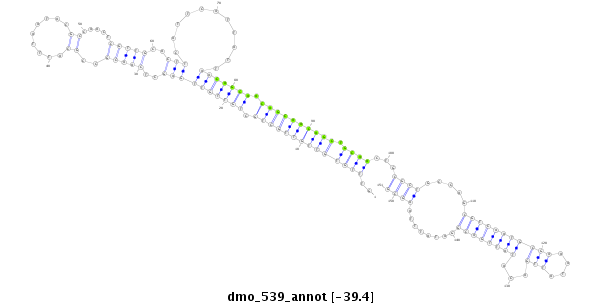 |
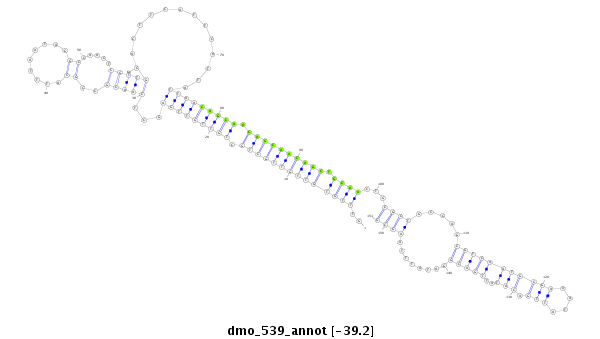 |
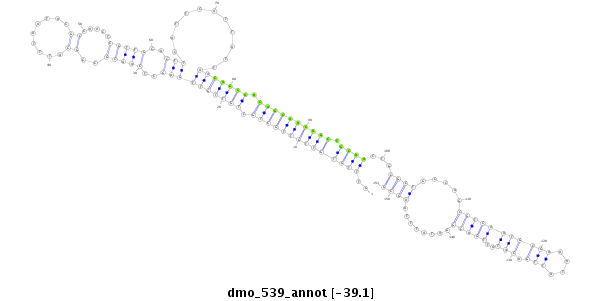 |
CDS [Dmoj\GI21621-cds]; exon [dmoj_GLEANR_6617:1]; intron [Dmoj\GI21621-in]
No Repeatable elements found
| mature | star |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AAAGCGGCGCAAGATCAAGAGCAATTGCCGGAGTCAGCTTTACGGTTCCTGTTTGTGTTGTTGCTGCTGTTGTTGAGCTCAACGCCGCATTTAATACGCCAACTCGTTGCACTTCATTCATTATTAACAGCAACAGCAACAGCTGCAACTGCGCTGCCAGCGTCAATGCGAAATATTGACATATTGACGGATATTTAAGCGAATGACTGAATGTGTGAGGCGAACTTAGATTGGCGGAGATATTTTATAGA **************************************************..((((((((((((((.(((((((((...(((((..((.........)).....)))))..............))))))))).)))))))))).))))...((((.....(((((((((((....)))...))))))))........))))************************************************** |
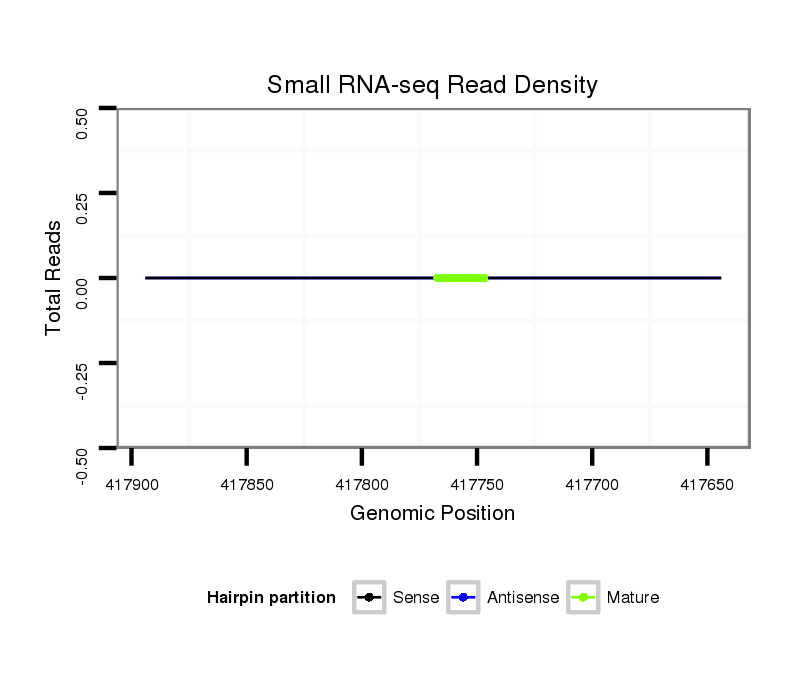
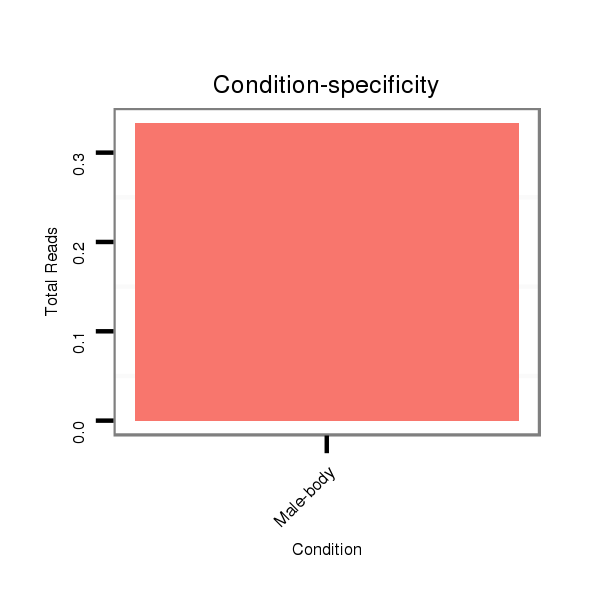
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V110 male body |
|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................................GATTGGTGGATCTATTTTAT... | 20 | 3 | 11 | 0.55 | 6 | 6 | 0 |
| ...............................................................................................................................CAGCAACAGCAACAGCTGCAA....................................................................................................... | 21 | 0 | 3 | 0.33 | 1 | 0 | 1 |
|
TTTCGCCGCGTTCTAGTTCTCGTTAACGGCCTCAGTCGAAATGCCAAGGACAAACACAACAACGACGACAACAACTCGAGTTGCGGCGTAAATTATGCGGTTGAGCAACGTGAAGTAAGTAATAATTGTCGTTGTCGTTGTCGACGTTGACGCGACGGTCGCAGTTACGCTTTATAACTGTATAACTGCCTATAAATTCGCTTACTGACTTACACACTCCGCTTGAATCTAACCGCCTCTATAAAATATCT
**************************************************..((((((((((((((.(((((((((...(((((..((.........)).....)))))..............))))))))).)))))))))).))))...((((.....(((((((((((....)))...))))))))........))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M060 embryo |
V110 male body |
M046 female body |
|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................CACAAATTCGCTTACTGTCTT........................................ | 21 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 |
| ............................................................................CTAGTAGCGGGGTAAATTA............................................................................................................................................................ | 19 | 3 | 17 | 0.06 | 1 | 0 | 1 | 0 |
| ..............................................AGGAGAAACTCAACAACG........................................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
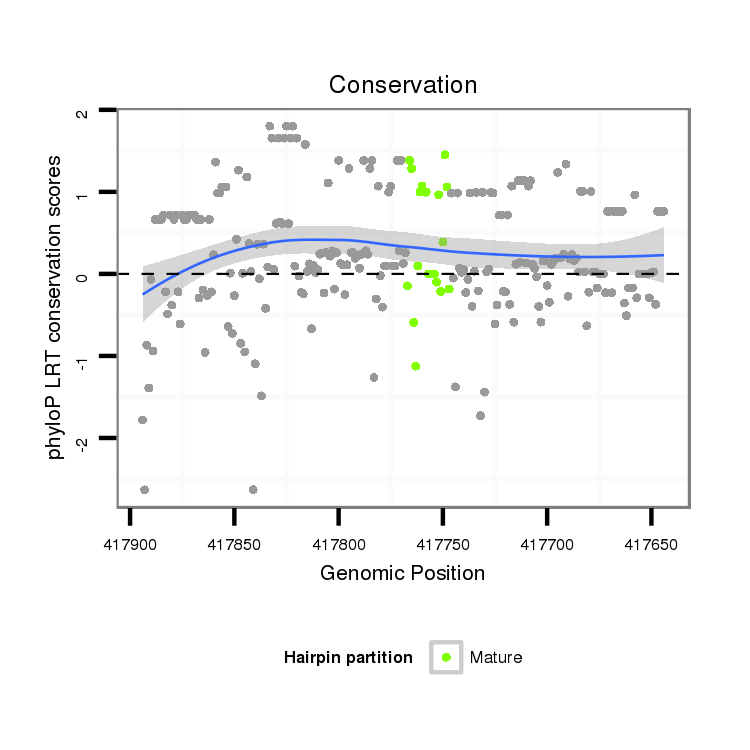
| droMoj3 | scaffold_6359:417644-417894 - | dmo_539 | AAAGCGGC-------GCAAGATCAAGAGCAATTGCCGGAGTCA-GCTTTACGGTTCCTGTTTGTGTTG---TTGCTGCTGTTGTTGAG-CT--CAACGCCGCATTTAATACGCCAACTCGTTGCACTTCATTCATTATTAACAGCAACAGCAACAGCTGCAACT----------------------------GCGCTGCCAGCGTCAATGCGAAATATTGACATATTGACGGATATTTAAGCGAATGACTGAATGTGTGAGGCGAACTTAGATTGGCGGAGATATTTTATAGA |
| droVir3 | scaffold_12928:3845256-3845461 + | AAACGGGC-------GCAGGATCAGCAGCAATTGCCGATGCCA-GCTTTGCTGT------TG-----TCCATTGTTGTTGTTGTTGAG-GT--CAGCGCCGCATTTAATACGCCAACTCGTTGCAGTT-GTTCATTATTAACAGCGACAGC------TGCAACT----------------TCGATGCCGACGTCGACGCCGGCGCCGACGCCAAATGTCGCCATATTGACAGTCATTAAAGCGAATAAATG------------------------------------------ | |
| droGri2 | scaffold_15081:3901507-3901742 - | AGATCAGCAGCAGCAGCAGCAGCAGCAGCAATTGACATTGCCAGGCTTTACTTT------TA-----T---TTGCTGTTGTTGTTGAG-GT--CAGCTTCGCATTTAATACGCCAACTCGTTGCAGCTCATTCATTATTAACAGGCACAGC------TGCAGC-AGTTGCAATGTTGACGTCGACCCCGACCCCGACACCGACACCGACGCCCAACGTTGCCATATTGACAGTTATTAAAGCGATTGAATGAATGAATGA--------------------------------- | |
| droWil2 | scf2_1100000004513:2022029-2022117 - | GTTACT----------------------------------------------G------TTGCTGTTG---TTGCTGTTGTTGTCGCT-CT--------TGTG-TTGTTGCTGCTGCTTATTACACATTGCCCACCATTGGTAGTGGCAGC------GGC------------------------------------------------------------------------------------------------------------------------------------- | |
| dp5 | 4_group3:5318454-5318496 - | AACAGC----------------------------------------------A------ACAGCAACA---------------------------------------------------------------------------------------ACAGCAGCA----------------------------ACAACAGCAGCAACAACG----------------------------------------------------------------------------------- | |
| droPer2 | scaffold_2:487472-487531 + | CGCACC-----------------------------------CA-GCTTTTAATTTTGCGTCTGTTTCG---CTGCCGTCGTCGTCGCT-CT---------------------------------------------------------------------------------------------------------------CACCGCCGC---------------------------------------------------------------------------------- | |
| droKik1 | scf7180000302592:198990-199102 - | ACG-------------------------------------------------GTTCCGGTAACTGTTG---TTGTTGTTGTTGTTGCCGCTTCAGCTGCCAGATGCCATGGCCCAAAGTGTCGCATT---------ACTGAAAGT----------GCGAC-------------------------------------------------------------------------AGTCACAGCGAATGCAAG---------------------------GG-----CACT---- | |
| droBia1 | scf7180000301754:4770757-4770830 - | GATACA-----------------------------------TA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GATACATAGATACATAGATACACAGATACATAGATACATAGA---TACATAGATGCATAGA-----TACATAGA | |
| droSec2 | scaffold_15:1293691-1293726 - | TGCT------------------------------------------------GCTGCTGTTTTTGCTG---CTGCTGTTGTTGTTGA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
Generated: 05/17/2015 at 10:33 PM