ID:dmo_532 |
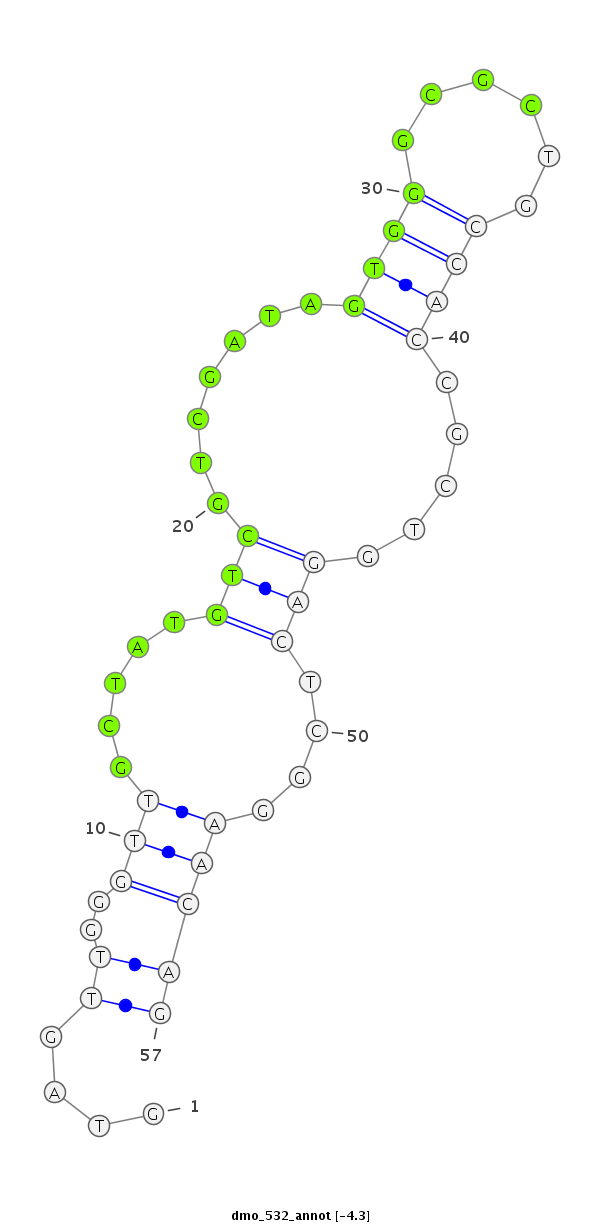
Coordinate:scaffold_6353:43437-43493 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
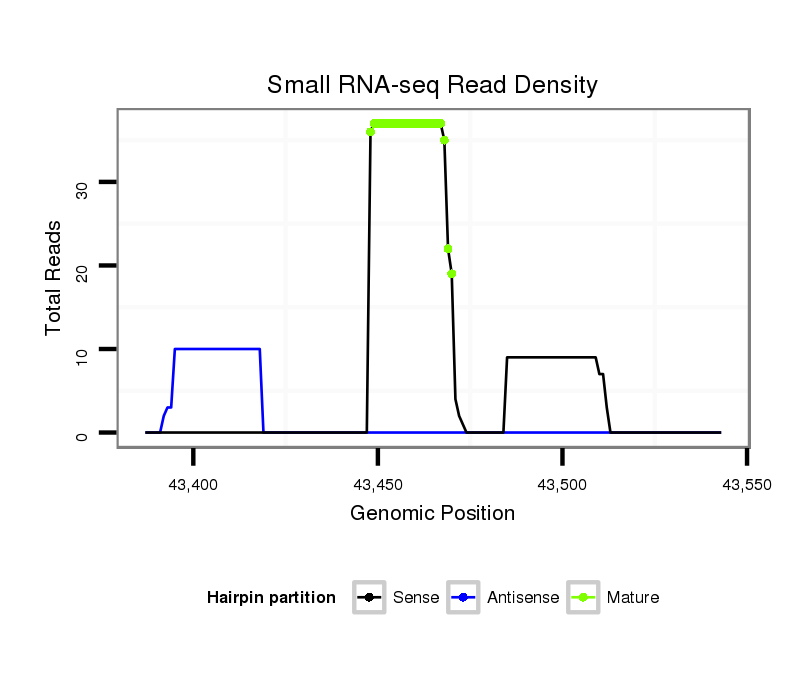
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
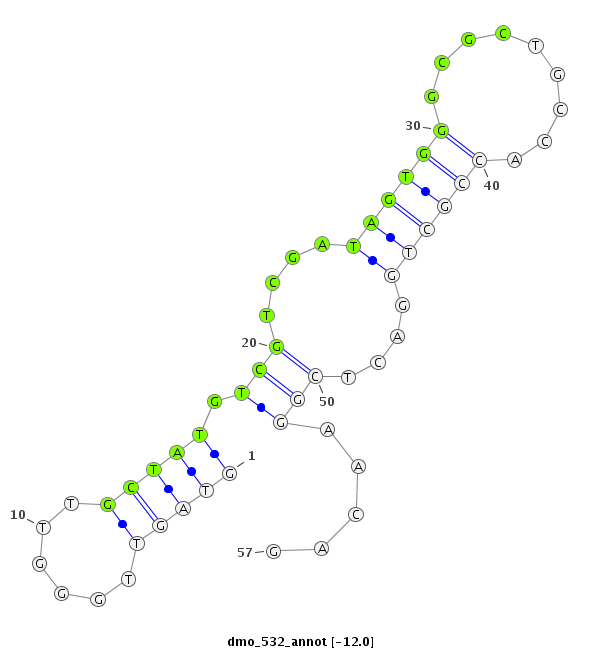
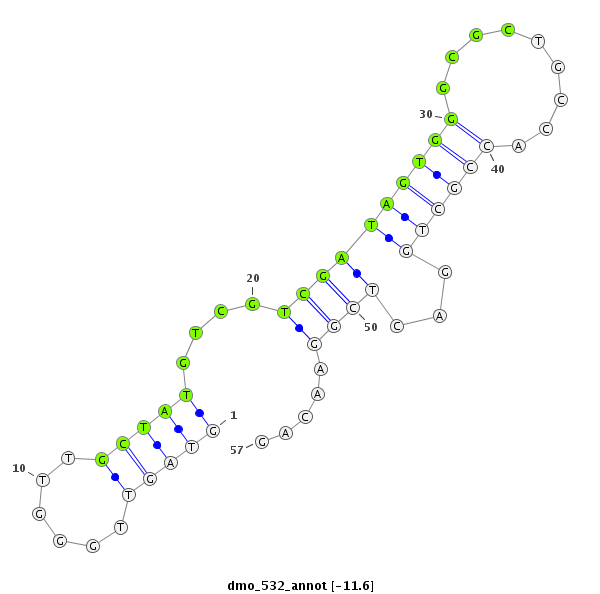
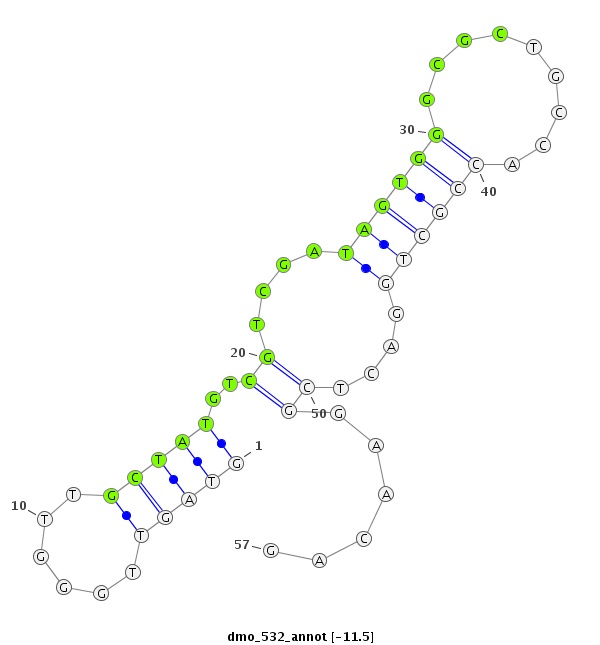
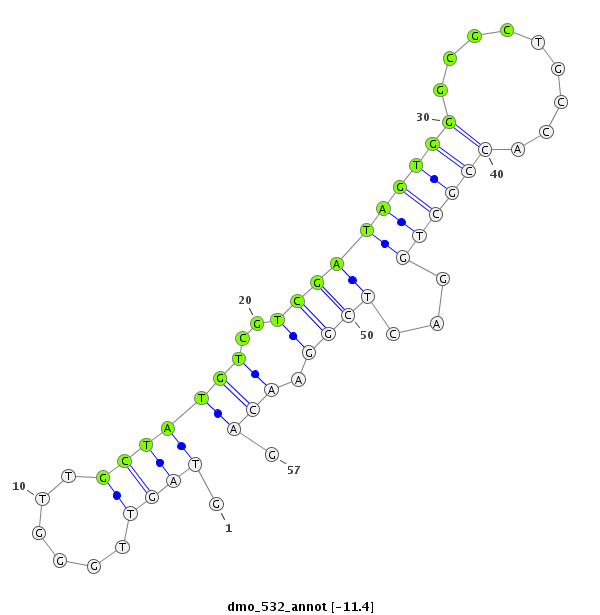
| -12.0 | -11.6 | -11.5 | -11.4 |
 |
 |
 |
 |
CDS [Dmoj\GI14561-cds]; exon [dmoj_GLEANR_15307:1]; CDS [Dmoj\GI14561-cds]; exon [dmoj_GLEANR_15307:2]; intron [Dmoj\GI14561-in]; Antisense to exon [dmoj_GLEANR_15305:1]; Antisense to CDS [Dmoj\GI14560-cds]
No Repeatable elements found
| mature | star |
| ##################################################---------------------------------------------------------################################################## ATACGCCAGGGCCGGATTGGCAACGATGGTCAGGGTGGCCACCCTCGCGAGTAGTTGGGTTGCTATGTCGTCGATAGTGGGCGCTGCCACCGCTGGACTCGGAACAGCAGAGTCACGCCGAGGGCCTCCGAGCGCTATACGCCGGAATCCCAAGGCG **************************************************....((..(((.....(((.......((((......)))).....)))....)))))************************************************** |
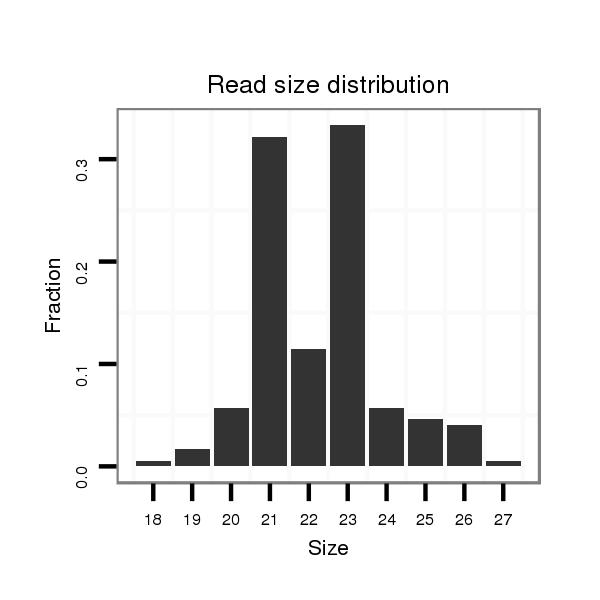
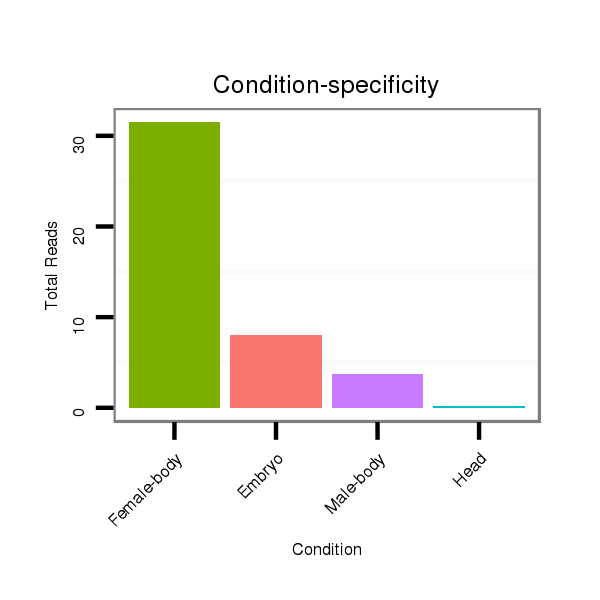
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V041 embryo |
V110 male body |
V049 head |
|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................GCTATGTCGTCGATAGTGGGCGC......................................................................... | 23 | 0 | 4 | 14.25 | 57 | 44 | 8 | 1 | 3 | 1 |
| .............................................................GCTATGTCGTCGATAGTGGGC........................................................................... | 21 | 0 | 4 | 13.75 | 55 | 24 | 4 | 17 | 10 | 0 |
| ..................................................................................................TCGGAACAGCAGAGTCACGCCGAGGGC................................ | 27 | 0 | 4 | 4.50 | 18 | 13 | 5 | 0 | 0 | 0 |
| .............................................................GCTATGTCGTCGATAGTGGGCG.......................................................................... | 22 | 0 | 4 | 3.50 | 14 | 14 | 0 | 0 | 0 | 0 |
| ..................................................................................................TCGGAACAGCAGAGTCACGCCGAGGGCC............................... | 28 | 0 | 4 | 3.25 | 13 | 4 | 9 | 0 | 0 | 0 |
| ..................................................................................................TCGGAACAGCAGAGTCACGCCGAGG.................................. | 25 | 0 | 4 | 2.50 | 10 | 9 | 1 | 0 | 0 | 0 |
| .............................................................GCTATGTCGTCGATAGTGGGCGCT........................................................................ | 24 | 0 | 4 | 2.50 | 10 | 8 | 1 | 0 | 1 | 0 |
| .............................................................GCTATGTCGTCGATAGTGGG............................................................................ | 20 | 0 | 4 | 2.00 | 8 | 7 | 0 | 0 | 1 | 0 |
| .............................................................GCTATGTCGTCGATAGTGGGCGCTGC...................................................................... | 26 | 0 | 4 | 1.75 | 7 | 7 | 0 | 0 | 0 | 0 |
| .............................................................GCTATGTCGTCGATAGTGGGCGCTG....................................................................... | 25 | 0 | 4 | 1.75 | 7 | 7 | 0 | 0 | 0 | 0 |
| ..............................................................CTATGTCGTCGATAGTGGGCGC......................................................................... | 22 | 0 | 4 | 1.25 | 5 | 5 | 0 | 0 | 0 | 0 |
| .............................................................GCTATGTCGTCGATAGTGG............................................................................. | 19 | 0 | 4 | 0.75 | 3 | 3 | 0 | 0 | 0 | 0 |
| .............................................................................................TGGACTCGGAACAGCAGAGTCACGCCG..................................... | 27 | 0 | 4 | 0.75 | 3 | 3 | 0 | 0 | 0 | 0 |
| .............................................................................................TGGACTCGGAACAGCAGAGTCACGCC...................................... | 26 | 0 | 4 | 0.50 | 2 | 1 | 1 | 0 | 0 | 0 |
| .............................................................GCTATCTCGTCGATAGTGGGCGCTGC...................................................................... | 26 | 1 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..................................................................................................TCGGAACAGCAGAGTCACGCCGAGGG................................. | 26 | 0 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..............................................................CTATGTCGTCGATAGTGGGC........................................................................... | 20 | 0 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...................................................................................................CGGAACAGCAGAGTCACGCCGAGGGC................................ | 26 | 0 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 |
| .............................................................................................TGGACTCGGAACAGCAGAGTCACGCCGAG................................... | 29 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..ACGCCAGGGCCGGATTGGCA....................................................................................................................................... | 20 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................GCTATGTCGTCGATAGTCGG............................................................................ | 20 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................GCTATGTCGTCGATAGTGGGCGT......................................................................... | 23 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................CTATATCGTCGATAGTGGGCG.......................................................................... | 21 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................CGGAACAGCAGAGTCACGCCAAGGGC................................ | 26 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................TTGACTCGGAACAGCAGAGTCACGCC...................................... | 26 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................TGGACTCGGAACAGCAGAGTCACGCCGT.................................... | 28 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......AGGGCCGGATTGGCAACGATGG................................................................................................................................ | 22 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................TCACGCCGAGGGCCTCCGAGCGCT..................... | 24 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................TCGGAACAGCAGAGTCACGCCG..................................... | 22 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................CAGCAGAGTCACGCCGAGGGCCTCCGAG......................... | 28 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................GCTATGTCGTCGATAGTGGTCGC......................................................................... | 23 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................TGCACTCGGAACAGCAGAGTCACGCCGAG................................... | 29 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................GCTATGTCGTCGATAGTGGGCGCTGCC..................................................................... | 27 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................GCTTTGTCGTCGATAGTGGGC........................................................................... | 21 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................TCGGAACAGCAGAGTCACGCCGAG................................... | 24 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................GCTATGTCGTCGATAGTGGGCC.......................................................................... | 22 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................AGCAGAGTCACGCCGAGGGCCTCCGA.......................... | 26 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................GCTACGTCGTCGATAGTGGGCG.......................................................................... | 22 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................TCGGAACAGCAGAGTTACGCCGAGGGCC............................... | 28 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................CGCTATGTCGTCGATAGTGGGC........................................................................... | 22 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................GCTATGTCGTCGCTAGTGGGG........................................................................... | 21 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................GCTATGTCGTCGATAGTG.............................................................................. | 18 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................TCGGAACAGCAGAGTCACGCCGAGGGCCT.............................. | 29 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................AGGGCCTCCGAGCGCTATACGCCGG............ | 25 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................CTATGTCGTCGATAGTGGGCGCT........................................................................ | 23 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................CTATGTCGTCGATAGTGGGCGCTGC...................................................................... | 25 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................GTCGTCGATAGTGGGCGCTGCC..................................................................... | 22 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................GGACTCGGAACAGCAGAGTCACGCC...................................... | 25 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................GCTATGTCGTCGTTAGAGGGCGC......................................................................... | 23 | 2 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................GCTATGTCGTCGATAGCGGGC........................................................................... | 21 | 1 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................TCGTCGATAGTGGGCGCTGCC..................................................................... | 21 | 0 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................CGGAACAGCAGAGTCACGCCGAGGGCC............................... | 27 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................TCTATGTCGTCGATAGTGG............................................................................. | 19 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................CACCTCTAGACTCGCAACAGC................................................. | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 |
| .................................................................TGTGGTCGACAGGGGGCGCT........................................................................ | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................AGCAGAGTCACGCAGTGG.................................. | 18 | 2 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................GTTGCTATGTCGTCTATGC................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ############################################################################################################################################################# TATGCGGTCCCGGCCTAACCGTTGCTACCAGTCCCACCGGTGGGAGCGCTCATCAACCCAACGATACAGCAGCTATCACCCGCGACGGTGGCGACCTGAGCCTTGTCGTCTCAGTGCGGCTCCCGGAGGCTCGCGATATGCGGCCTTAGGGTTCCGC **************************************************....((..(((.....(((.......((((......)))).....)))....)))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V110 male body |
V041 embryo |
V056 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ........CCCGGCCTAACCGTTGCTACCAGT............................................................................................................................. | 24 | 0 | 4 | 7.00 | 28 | 10 | 17 | 1 | 0 | 0 |
| .....GGTCCCGGCCTAACCGTTGCTACCAGT............................................................................................................................. | 27 | 0 | 4 | 2.00 | 8 | 8 | 0 | 0 | 0 | 0 |
| ......GTCCCGGCCTAACCGTTGCTACCAGT............................................................................................................................. | 26 | 0 | 4 | 1.75 | 7 | 5 | 1 | 0 | 1 | 0 |
| ........CCCGGCCTAACCGTTGCTACCATT............................................................................................................................. | 24 | 1 | 4 | 1.00 | 4 | 0 | 4 | 0 | 0 | 0 |
| .......TCCCGGCCTAACCGTTGCTACCAGT............................................................................................................................. | 25 | 0 | 4 | 0.75 | 3 | 3 | 0 | 0 | 0 | 0 |
| ....CGGTCCCGGCCTAACCGTTGCTACCAGT............................................................................................................................. | 28 | 0 | 4 | 0.75 | 3 | 2 | 1 | 0 | 0 | 0 |
| ..........................................GGAGCGCTCATCAACCCAACGATACAGGA...................................................................................... | 29 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................GAGCGCTCATCAACCCAACGATACAGCA...................................................................................... | 28 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................TGAGCCTTGTCGTCTCAGTGCGGCT.................................... | 25 | 0 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 |
| .............................................GCGCTCATCAACCCAACGATACAGC....................................................................................... | 25 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........CCGGCCTAACCGTTGCTACCAGT............................................................................................................................. | 23 | 0 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 |
| .............................................................................................................................GAGGCTCGCGATATGCGGCCTTAGGGT..... | 27 | 0 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................CACGGTGGCGACCTGAGCCTTGTCGT................................................ | 26 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................CGGAGGCTCGCGATATGCGGCCTT.......... | 24 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........CCCGGCCTAACCGTTGCTACTAGT............................................................................................................................. | 24 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................GGCGACCTGAGCCTTGTCGTCTCAGTG......................................... | 27 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................AGGCTCGCGATATGCGGCCTTAGGGT..... | 26 | 0 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| .........CCGGCCTAACCGTTGCTACCATT............................................................................................................................. | 23 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....GGTCCCGGCCTAACCGTTGCTACCAGTC............................................................................................................................ | 28 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....GGTCCCGGCCTAACCGTTGCTACCA............................................................................................................................... | 25 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................GGCGACCTGAGCCTTGTCGTCTCAGT.......................................... | 26 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................CACCCGCGACGGTGGCGACCT............................................................ | 21 | 0 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................CTGAGCCTTGTCGTCTCAGTGCGGCT.................................... | 26 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................CCTGAGCCTTGTCGTCTCAGTGCGGCT.................................... | 27 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........CCCGGCCTAACCGTTGCTACCAGTCCC.......................................................................................................................... | 27 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................CCGTTGCTACCAGTCCCA......................................................................................................................... | 18 | 0 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......GTCCCGGCCTAACCGTTGCTAACAGT............................................................................................................................. | 26 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....GGTCCCGGCCTAACCGTTGCTACC................................................................................................................................ | 24 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......TCCCGGCCTAACCGTTGCTACCAG.............................................................................................................................. | 24 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................TGGCGATCTGAGCGTTTTCG................................................. | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...........CCCCTAACCGTTGCTACCAGT............................................................................................................................. | 21 | 2 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........GGCCTAACCGTTGCTACCAG.............................................................................................................................. | 20 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................CCGGTGGAAGCGCTTATCG...................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................CGGTGGAAGCGCTTATCG...................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6353:43387-43543 + | dmo_532 | ATACGCCAGGGCCGGATTGGCAACGATGGTCAGGGTGGCCACCCTCGCGAGTAGTTGGGTTGCTATGTCGTCGATAGTGGGCGCTGCCACCGCTGGACTCGGAACAGCAGAGTCACGCCGAGGGCCTCCGAGCGCTATACGCCGGAATCCCAAGGCG |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
Generated: 05/17/2015 at 10:31 PM