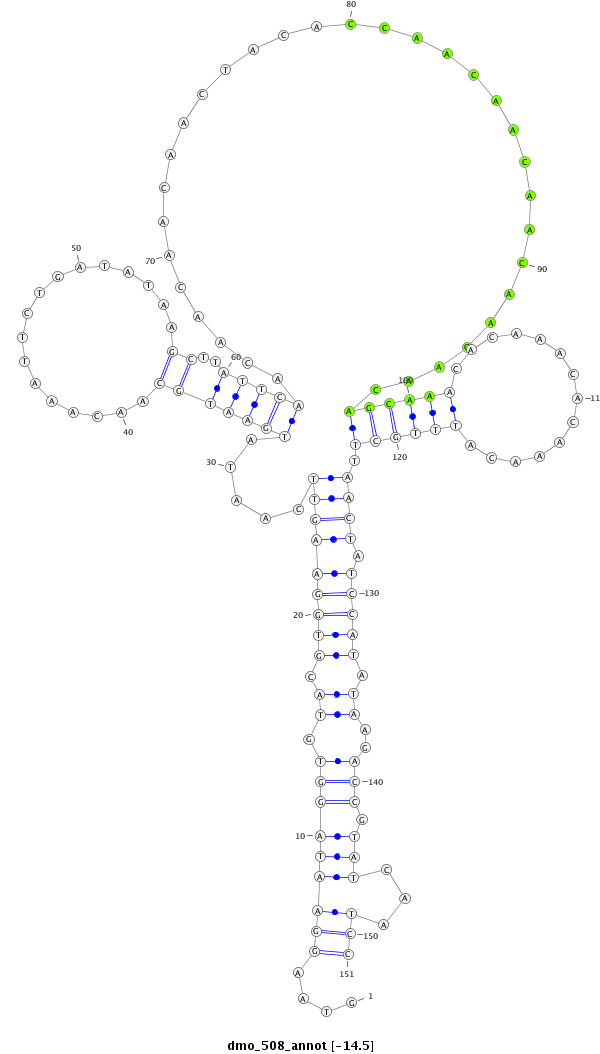
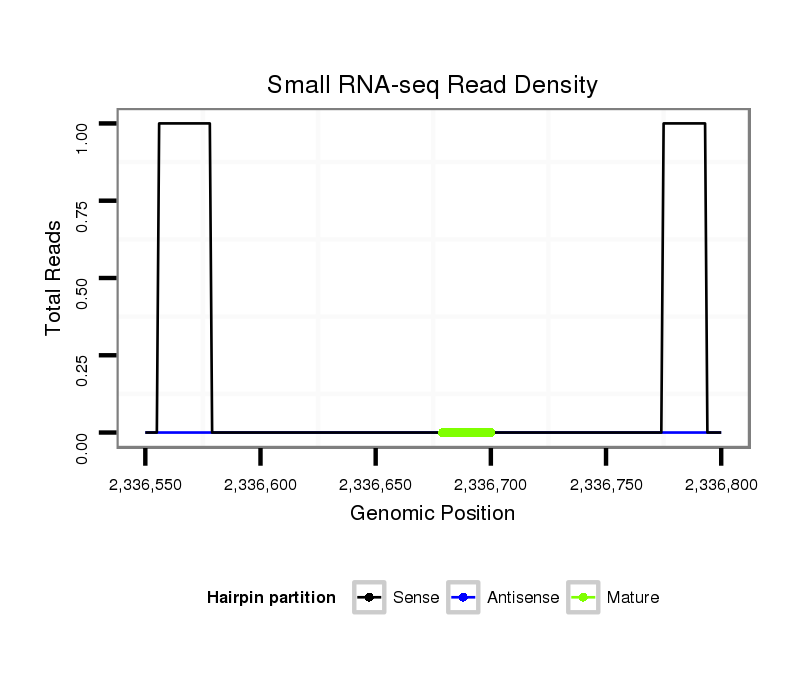
| droMoj3 |
scaffold_6328:2336550-2336800 + |
dmo_508 |
AGGATGGC-----GT-GG-------------------------CCATCGATGTCG------CTGG---------------------------------------------CGGCCAA-----------------AT-------------------------ATATGAG-----------CAG------GAG--------------------GTAA-----------------------------------------GGAATAGGTGT-----------------------------ACGTGGAAGTTCAATATGAA------------------------------------------------------------TGCAACAAATTC------TGATATA-A------------------------------------------------------------------------------------------G------------------------------CTTATTCAACAACAACAA---CTACACCAACA---------------------ACAACAACAACA------GCAAACAC-AAACA---------CAAACATTTGCTTAACT---------------------------------------------------------AT--------------------------------CCAT----AT-------AAGACCGTATCAATCCATTTGTCAATA----TT-----GGTTGAGGGCCTG----------------------------------------------------------------AACT-GACTGCCAAAGACATAATCA |
| droVir3 |
scaffold_13246:2518156-2518354 - |
|
AAGCTAGCA----AG-AT--------G----------------CCCGCCACGGTG------CT--------------------------------------------------------------------------------------------------------------------------------------------------------------GGCTGCTGCCTC--------------------------------------------------CGGT--------------------------------------------------------------------CACA------------------------CGAC------CAAGC-TAAGC------------------------AACAACAAC-------------------------------------------------------------------------------------------------AACAACAA---CAATAGC---------------------------AGCAACAATAGCAACAA------C-AAACACA----ACAA---------------------------------------------------------------------------T----ATAATGCTTAACAACAACATGAACAA-TAGCTTATGC------A------ACTCGATCGCCCATCTAAGCGAT------------TCAAGGACCA----------------------------------------------------------------ATCT-CACTGTCA-----------A |
| droGri2 |
scaffold_15252:8165775-8166046 - |
|
AGCGTCAGA----GC-AAACAGAGA-G----------------AAAGCGACAG-GCTGGCAAATGCCAGGGACGACATTGTGGACGAGAAATCCTCGTGGTT--GTT--------------------------------------------------------------TGA------------------------TTC--------------------------------------------------------------GAAAATTGAGTTTTAGCGGCGCAAATAAA-----------------------------------------------------------------------------------------------AAT------AAAACTT-AGAATACCAACAGCAACAGCAACAGC---------AGCAGCAGCA---ACAG----------------------------------------------------------------------------------------------------------------------------CAACAGCAGCA------GCAAAAAC-------A----ACAACAACAAAAGCTCAACC---------------------------------------------------------AT--------------------------------TAAA-------------AAGGAGTTATCACCCAATTT---AGCACCA-C----A-AGCG-------CG----------------------------------------------------------------AAAT-AACGCACACAAACACAGTCA |
| droWil2 |
scf2_1100000004511:5742606-5742831 - |
|
AGTGTAGGA----GT-TGTTGTAG-------------------TCGCC---------------------------------------------------------------GGTC----------------------------------------------------------------------------------------------------------------------------------------------------------------ATCATCATCAGC---------ATCAGCAACATCAGCATCAG---------------------------------------------------------------------------------------------------------------------------------CA---ACATCAACACCACAACCAACATCAACAGC------AACAACA----------------------------------------A------CAA------------CAA------------------------CAGCAACA------A------C-AAATGCAAATGGTGG--------GCACAACTAGCTCCA----------------------TGGATG-----------------------------------------------------------------------------------------------------------------------------------------GCACCAGCCAGGTGGCCATGTGTCATGATGGTTTAGTTGGCACAGCAAATGAACT-GACTGTCTATGATGTGATTA |
| dp5 |
3:5866323-5866541 - |
|
AAAGATACA----AC-AA-------------------------CAGCTGT---TG------CT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCAACAGCCACAAATACTGCCCCAACAACAGCCACAAA-----------------------------------------------------------------------TA------CTGCC-------------------------------------AC---AACAACAACAACAA----------------------------------------------------------------------------------------------------------------CAAAAGCTA------CAACAACAACAACAA------C-A----------AC-----------------AGGCGTTG----------------------TGGGCGGCATTAGGCGTGGCTCGAGTGC-------AAAATGCAAAACAACATTGAAGACGAGAAAATCTGGCTACTGTA------ATTTGT---------------------------------------------------------------------------------------------------------CT-GCCCACCAAAGACA---CGA |
| droPer2 |
scaffold_12:1683222-1683441 + |
|
AGCAGCAG-----CT---------------------------------GGTGGCC------CAGGCGGCGGACGGCAGCGAGAGTGGGGCGCC---GATGTC--GAT--------------------------------------------------------------TGC-----------------------------------------------------------------------------------------------------------------CCAGGTGCAGACCCTGCAGGGGCATCAGA-T---------------------------------------CATAG------GCA-ACTTGAATCAGGTGAATATGACTG----------------------------------ATTTCCAAC-------------------------------------------------------------------------------------------------AACAGCAG---CCACAGCAGCA---------------------GCCACCACAACA------GCAAACAC-------A----ACCCCAACAGACGCTC-------------------------------------------------------------------------------------------------------------TGA-------TCG------------------------GACA------AGACCGA----------------------------------------------------------------AACGTAGAATCTA----------TT |
| droAna3 |
scaffold_13337:10557351-10557574 - |
|
AACTTCCAT----AT-GG-------------------------GCATCAGTG----------------------------------------------------------------------------------------------------------------ACGATGGCTCGGAGACT-----------------------------------------------------CACAGTGAAGACAACAAGCTG--------AT------GCACAGCAACAGCAGCAATC-----AGATGCAGCAACATCAGC---------------------------------------------------------------------------------------------------------------------------AGCAGCAGCAGCAAATG------------ATG---CAGCAAC------AACAGCA----------------------------------------G------CAG------------------------------------------CAACAGCAACAG------C-A----------GT---CC---------AA---------------------------------------------------------------------------------------------TAACTTCTTT------A------GCTCCAATCACTTTCTCAACA----ATAGTCAGAATCAGAACCAA----------------------------------------------------------------AATC-AGTTGCCAAA---------- |
| droBip1 |
scf7180000396691:631385-631625 - |
|
ATAACCGC-----ATCGCC-ATCGTCATCGAAAAGCGAAGCGGCCA----------------------------------------------------------------------C-----------------AAACAATAA-----------------------------------------------------------------------------------------------------------------------------------------CAG------------------------------CAGCAGCAGCAGCACAAGCGGCACTAGCATAGTCGT---------AGCCACTAACGGTATCAAAGAGAGTAACAGTAACTT----------------------------------AAGTACAAC-------------------------------------------------------AA------------------------------CAGCACCGTCAG---CAACGCCAGCAT---CA------------TCAGCCA------CACCAGCAACAACAA------C-A----------AC---CA---------TACC------A-----TATCTGGTCGGCG-----------------------------------------------------------------------------------------------TTCCAACC---AGCAGTGA-------GGTG-AG-----------------------------------------------------------------------------TT-------GTGTGATCC |
| droKik1 |
scf7180000302391:1564221-1564399 + |
|
AACATCAAC----AC-AG-------------------------CAACTAATAAAA------CAA----------------------------------------------------------------------------------------------------------AGGCCAAAGCTTAAGGAGGAA--------------------ACAAAAACAACTTCTGTAAGACATAAAACAATAATACCACAGCTTTAAACC------------------------GATAT-----TTATGC------------------------------------------------------------------------------------------------------------------------------AATTGCAAC-------------------------------------------------------------------------------------------------ACCAGGAA---GAACATCAACA---------------------GCTAGATCAATA------ACGAAATGGAGACA---------CCAACACT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TACCTA |
| droFic1 |
scf7180000454065:89154-89378 - |
|
AACTTTCAC----AT-GG-------------------------GCATCAGCG----------------------------------------------------------------------------------------------------------------ACGATGGCTCCGAGACC-----------------------------------------------------CAGAGCGAGGACAACAAGATG--------AT------GCACAGCTCCAGCAGCAATC-----AAATGCAACAGCAACAGC---------------------------------------------------------------------------------------------------------------------------AGCAGCAGCA---ACAT------------CAG---CAAC----------------AT------------------------------CAGCAGCAGCAGCAG---CAG---------------------CAAATCCTGCA------GCAACACCAGCAG------C-A----------GT---CA---------AA---------------------------------------------------------------------------------------------CAACTTCTTC------A------GTGCGAATCCCTTTCTCAGCA----ACAGCCAGAACCAGAACCA-----------------------------------------------------------------------------------------A |
| droEle1 |
scf7180000490546:821347-821584 - |
|
ATGATAATG----AC-GA-------------------------CAACAGGACGTG------AACCTGGTGAACTAGAACTAGAA-----------T----CAAGACAGCC-AATC--------------------------------------------------------------------------------------------------------------A---------------------------------------------CTTTAGCAACTGCAACAAG-----------------------------------------------------------------------------------------------TAC------AAATCTA-ACCATAACAACAACAACAACAAC---------------GACAGCAAC--------------AACTAGCAACAACAAC------AACAACA----------------------------------------G------CAA------------CAACAGCAACAACAATTCCAACG-----------------------------------------------------CAAC---------------------TGGCAGTAC------------------------------T-----------------GCAA----------CAAC--------------------TCCAAGTCCAGAACTAGACA-------ACCAGGTG-GAGAC------------------------------------------------------------------------------------CTTAACCA |
| droRho1 |
scf7180000779841:473239-473452 - |
|
AGCAACAGC----AGCAAC-ACAATCAGCAGCAACAGATGCAACAA----------------------------------------------------------------------C-----------------A-----------------------------------------------------------------------------------------------------------------------G--------AT------T--CAACAACAGCAGCAA-C-----AGATGCAACAACAGCAGC---------------------------------------------------------------------------------------------------------------------------A------------ACAGCAGCAACACAATCATCAGCAACAGCAGCAAC-------TTAATCATCAGCAACAGCAGCAACTTAATCATCAGCAAC---AGCAGCAA---CACAATCATCA---------------------GCAACAGCAGCA------ACTTAATC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AACAG-CAGCAACACAATCA |
| droBia1 |
scf7180000302402:3602359-3602525 - |
|
GAGTGGCCA----GC-AA-------------------------ATACCAGAG-------------------------------------------C----CGAAATCTAT-GCGAAA-----CC----------------------CAGCCAGGGGCAACAACACAAA-----------CCTACCTGGCCAGTAAAAACAGCCACGAAGAAG------------------------------------------TGGAAACGAATAT-----------------------------ACATAA------------------------------------------------------------------------------------------------------------------------------AAATCAAGT---------------------------------ACCAGCAGC------AGCAGCA----------------------------------------G------CAA------------CAA------------------------CAGCAACA-------------C-AAACA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAAACA |
| droTak1 |
scf7180000415211:49430-49586 - |
|
CGAATGTGGTTCAGC-GG-------------------------CAT--------------------------------------------------------------------------------------------------------------------------------------------------T----------------------------TGTCG---------------------------------------------CCCCATCACCAGCAGCAGTCGCCGCACCTGCAACAGCAACAGCAG---------------------------------------------------------------------------------------------------------------------------CAGCAGCA---ACAC------------CTA---CACC----------------AG------------------------------CAGCAGCAGCAGCAA---CACCTACACC---A------------------GCAACACCAC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGCAGCAGCAGCAA----------------------------------------------------------------CCAC-AGGCGCTGGC---------- |
| droEug1 |
scf7180000409672:210090-210284 + |
|
TGGCATC-----------------------------------------GGTGGCG------CAGG----------------------------------------------------------------------------------------CGGCAGCAGTACGAG-----------CAGGACGAGGTG---------------------------TACGTCCGCCGCCTG--------------------------------------------------TGGC--------------------------------------------------------------------CATGCC----------------------CATT------CTGGG-TGTGC------------------------AATTGCAAC-------------------------------------------------------------------------------------------------AACAGGAA---CTGCAGC---------------------------------AGCAACG-CGG------C-G----------G------------------------------------------------------------------------------TCTGC-CAGTGCCCAGCCAAGATATATCTAG-TAGCTC--------AGG-------TCCGACAAGTCG--------------------------------------C--CA----------------------------------------------------------AAATGCAGCCAAAGTAGTTT |
| dm3 |
chr3L:7757404-7757651 - |
|
TGCATGCAG----AT-GG-------------------------CCACCGAA--------------------------------------------T----CGAGTCCTTC-GATCAA-----CT----------------------TTCACATGGGCATCAGCGATGATGGCTCCGAAACG-----------------------------------------------------CAGAGCGAGGACAACAAGATG--------AT------GCACACTTCCGGCAGCAATC-----TGGTGCAGCAGCAACAGC---------------------------------------------------------------------------------------------------------------------------AGCAGCAGCAACAACAA----------------------------------------------------------------------------------------------------------------CAAATCCTGCA------GCAGCATCAACAG------C-A----------GT---CA---------AA---------------------------------------------------------------------------------------------TAGCTTCTTC------A------GTTCGAATCCCTTTCTCAACA-------GTCAGAATCAGAATCAA----------------------------------------------------------------AACC-AATTGCCAAA---------- |
| droSim2 |
2r:5176665-5176812 - |
|
AGAAAGTC-----GT-GG-------------------------CCAACAATA---------------------------------------------------------------------------------------ATAAAATCAATCAGCGGCAACATGGCCAG-----------CCGGCCGTGGCATT----------------------------TGCCATC--------------------------------------------------------------------------------------------------------------------------------------------------ACATCGACTAACAATCACAT----------------------------------AAGCGGCC------------------------------------CAGCAGC------AACCATA----------------------------------------A------CAA------------CA---------------------A---CAGCGGCA------ACTGGCGA-------A----ACAACAACAG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A |
| droSec2 |
scaffold_0:71055-71299 - |
|
TACATGCAG----AT-GG-------------------------CCACCGAA--------------------------------------------T----CGAGTCCTTC-GATCAA-----CT----------------------TTCACATGGGCATCAGCGATGATGGCTCCGAAACG-----------------------------------------------------CAGAGCGAGGACAACAAGATG--------AT------GCACACTTCCGGCAGCAATC-----TGGTGCAGCAGCAACAGC---------------------------------------------------------------------------------------------------------------------------AGCAGCAACA---ACAA----------------------------------------------------------------------------------------------------------------CAAATCCTGCA------GCAGCATCAACAG------C-A----------GT---CA---------AA---------------------------------------------------------------------------------------------CAGCTTCTTC------A------GTTCGAATCCCTTTCTCAACA-------GTCAGAATCAGAACCAA----------------------------------------------------------------AACC-AGTTGCCAAA---------- |
| droYak3 |
X:389187-389403 + |
|
AGATGGGAG----AG-GA-------------------------GAAATGATT------------------------------------------------TT--TTTTT-CGTCAAAAGGAGCGCAGTTTCAATA-------------------AGCAA-----------GAATGGAAGGGTAGAAGAAGA--------------------AGAAAGATAAATTCAATAAATGGTAATATATCTATATATT--------------------------------------------------------------------------------------------------------------------------------------------------------A------------------------AAT-------------------------------------------------------------------------------------------------------TAAAGCAA---GCAAGCAAGCA---------------------GAAA----GGCA------A------C-A----------A------------------------------------------------------------------------------TATAA----AGC-------------AAGCAATTAAATTTAAG------T------ATTTGAATGTTTA--------------------------------------A--CA----------------------------------------------------------GAACGCCGTAAACATCCTTT |
| droEre2 |
scaffold_4770:7639861-7640117 - |
|
ACGAAACA-----AC-GG-------------------------CCACTAACA---------------------------------------------------------------------------------------ACAA-----------------------------------------------------------------------------------------------------------------------------------------TAGCAAC---------ACAAGCAGCAGCAGCAGCAG------------TAGCAGCAACAACATAGTCATACCGGCATCGGCCACTAACGGTATCAAAGAGAGTAACAGTAACTT----------------------------------AAGTACAAC-------------------------------------------------------------------------------------------------AACAGCAA---CAGTAGCAGCA---------------------GCAACAACAGTA------GAAGGAGT--A---------GCACCAGCAATAAC--------CTCCACAATCGTGGTGACCGGCGGCAC------------------------------T-----------------------------------------------------------CCTCCATTG---AGCAGTTT-------GGTG-AG----TAGCTCTGCCCCAAAAC------------------------------------------------------------------ACCAACCA |