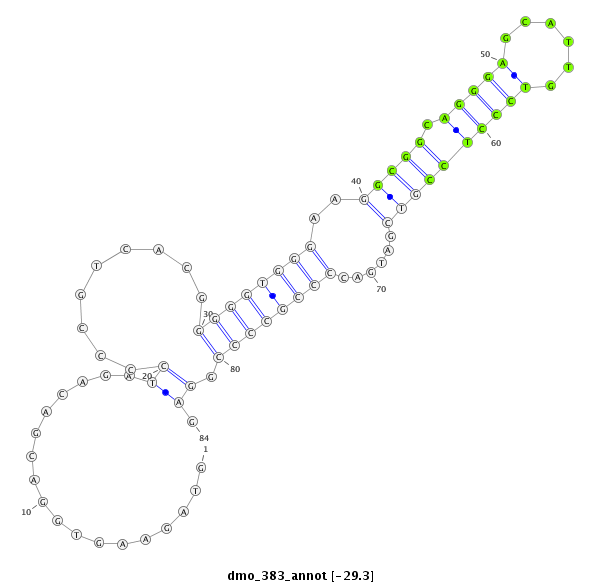
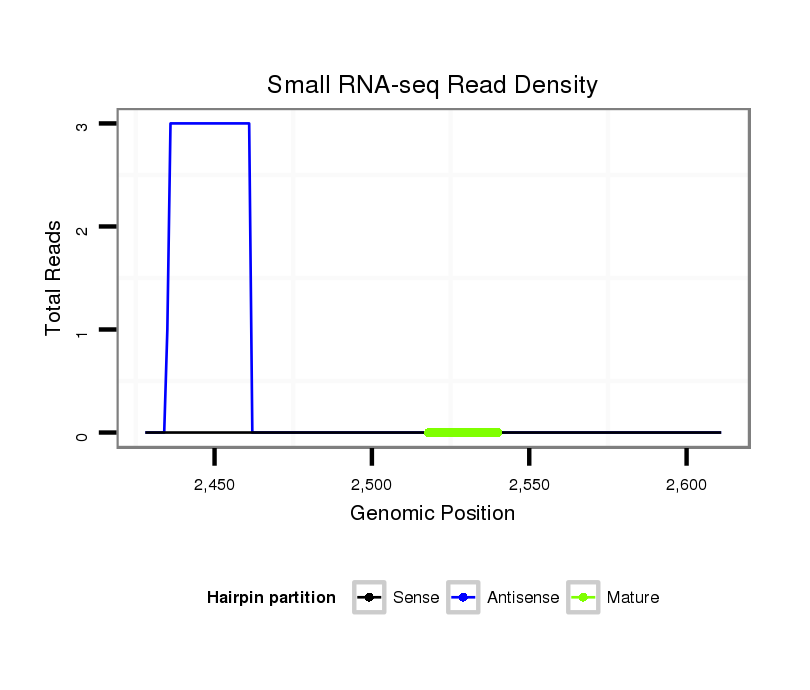
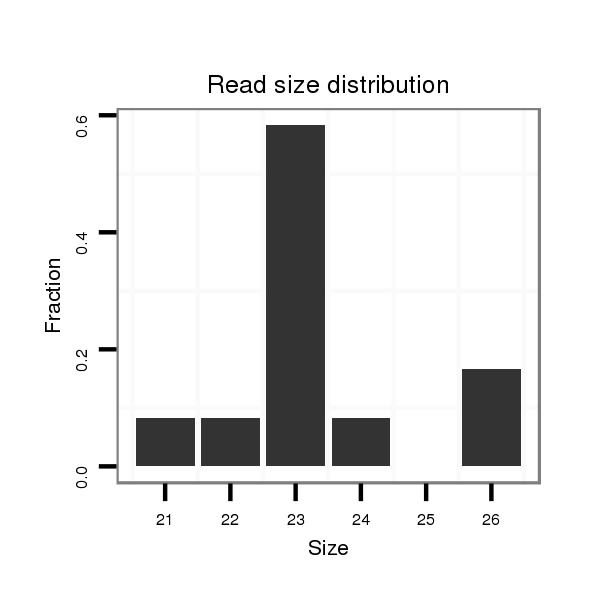
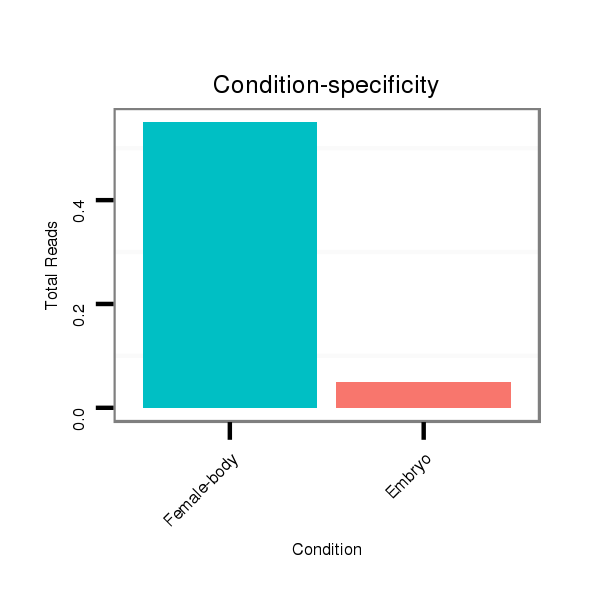
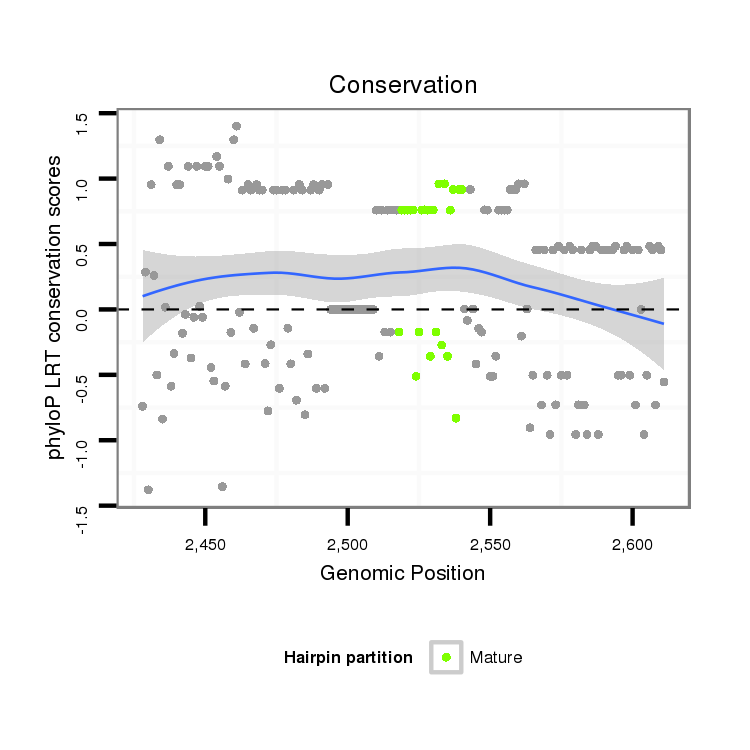
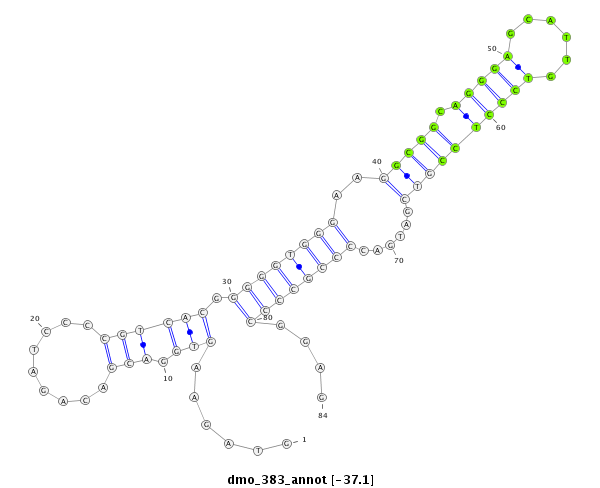
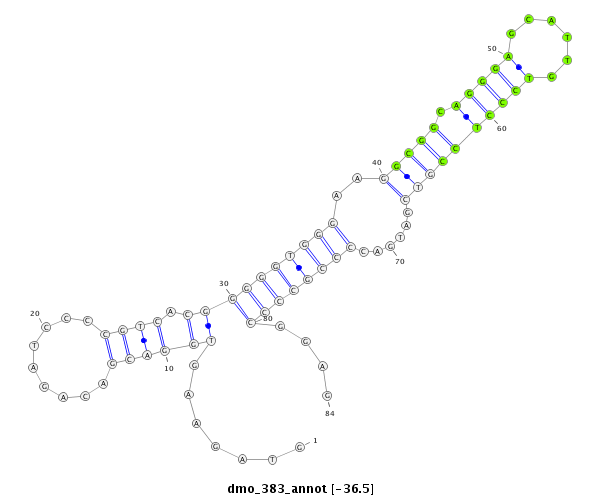
ID:dmo_383 |
Coordinate:scaffold_5676:2478-2561 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -37.1 | -37.1 | -36.9 | -36.5 |
 |
 |
 |
 |
CDS [Dmoj\GI18321-cds]; exon [dmoj_GLEANR_3225:1]; CDS [Dmoj\GI18321-cds]; exon [dmoj_GLEANR_3225:2]; intron [Dmoj\GI18321-in]
No Repeatable elements found
| ##################################################------------------------------------------------------------------------------------################################################## GGGCGACTCCGACGCCGTGGACGGCAAGCGAGGACGAAGAAGCTCCGGCGGTAGAAGTGGACGACAGATCCCCGTCACGGGGGTGGGAAGGCGGCAGGGAGCATTGTCCCTCCGTCGATGACCCCGCCCCGGAGACCTGGCGCGGGATGGAGCGGAGGAAGGGCGCAGTGATGTGTCGAGAAGT **************************************************..................((.........((((((((..(((((.(((((......))))))))))......)))))))).)).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V110 male body |
V041 embryo |
V049 head |
V056 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................CAAGCGAGGACGAAGAAGCTCC.......................................................................................................................................... | 22 | 0 | 10 | 0.90 | 9 | 8 | 1 | 0 | 0 | 0 | 0 |
| ........................CAAGCGAGGACGAAGAAGCTCCG......................................................................................................................................... | 23 | 0 | 10 | 0.80 | 8 | 7 | 0 | 1 | 0 | 0 | 0 |
| ........................CAAGCGAGGACGAAGAAGCTCCGG........................................................................................................................................ | 24 | 0 | 10 | 0.60 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................GCGGCAGGGAGCATTGTCCCTCC....................................................................... | 23 | 0 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 | 0 | 0 |
| ........................CAAGCGAGGACGAAGAAGCTCCGGC....................................................................................................................................... | 25 | 0 | 7 | 0.29 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................GGCGCAGTGATGTGTCGAGC... | 20 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................AAGCGAGGACGAAGAAGCTCCG......................................................................................................................................... | 22 | 0 | 10 | 0.20 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ........................CAAGCGAGGACGAAGAAGCTC........................................................................................................................................... | 21 | 0 | 10 | 0.20 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ........................CAAGCGAGGACGAAGAAGC............................................................................................................................................. | 19 | 0 | 12 | 0.17 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................GAGGACGAAGAAGCTCCGGC....................................................................................................................................... | 20 | 0 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................CGAGGACGAAGAAGCTCCGGC....................................................................................................................................... | 21 | 0 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................AAGCGAGGACGAAGAAGCTCCGGCGG..................................................................................................................................... | 26 | 0 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................CGAGGACGAAGAAGCTCCGGCGG..................................................................................................................................... | 23 | 0 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................AAGCGAGGACGAACAAGCTCCGG........................................................................................................................................ | 23 | 1 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................CAAGCGAGGACGAAGAAGCTCCGT........................................................................................................................................ | 24 | 1 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...CGACTCCGACGCCGTGGACGG................................................................................................................................................................ | 21 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................CAGGGAGCATTGTCCCTCCGT..................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................GCGGCAGGGAGCATTGTCCCTCA....................................................................... | 23 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................GGGGGAGGGAAGGGGGCATGGAG................................................................................... | 23 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................GCGGCAGGGAGCATTGTCCCTCCGTC.................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................TGGGAAGGCGGCAGGGAGCATTGTCC........................................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................CCTCGTCCAGGGGGTGGGA................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................ATTGTGCCTCCGTCGGTCA............................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................GCGGCAGGGAGCATTGTCCCTCCG...................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................GCGGCAGGGAGCATTGTCCCTC........................................................................ | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................GACCCCCCCCGGGAGACCA.............................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
|
CCCGCTGAGGCTGCGGCACCTGCCGTTCGCTCCTGCTTCTTCGAGGCCGCCATCTTCACCTGCTGTCTAGGGGCAGTGCCCCCACCCTTCCGCCGTCCCTCGTAACAGGGAGGCAGCTACTGGGGCGGGGCCTCTGGACCGCGCCCTACCTCGCCTCCTTCCCGCGTCACTACACAGCTCTTCA
**************************************************..................((.........((((((((..(((((.(((((......))))))))))......)))))))).)).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V041 embryo |
V110 male body |
V056 head |
V049 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ........GGCTGCGGCACCTGCCGTTCGCTCCT...................................................................................................................................................... | 26 | 0 | 11 | 2.55 | 28 | 23 | 1 | 2 | 1 | 1 | 0 |
| .......AGGCTGCGGCACCTGCCGTTCGCTCCT...................................................................................................................................................... | 27 | 0 | 11 | 1.18 | 13 | 13 | 0 | 0 | 0 | 0 | 0 |
| .........GCTGCGGCACCTGCCGTTCGCTCCT...................................................................................................................................................... | 25 | 0 | 11 | 0.82 | 9 | 8 | 1 | 0 | 0 | 0 | 0 |
| ...............................CCTGCTTCTTCGAGGCCGCCATCTT................................................................................................................................ | 25 | 0 | 7 | 0.71 | 5 | 1 | 2 | 1 | 1 | 0 | 0 |
| ......GAGGCTGCGGCACCTGCCGTTCGCTCCT...................................................................................................................................................... | 28 | 0 | 11 | 0.55 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| ..........CTGCGGCACCTGCCGTTCGCTCCT...................................................................................................................................................... | 24 | 0 | 11 | 0.36 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ............GCGGCACCTGCCGTTCGCTCCTGCTT.................................................................................................................................................. | 26 | 0 | 11 | 0.27 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ...........TGCGGCACCTGCCGTTCGCTCCTGCTT.................................................................................................................................................. | 27 | 0 | 11 | 0.27 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| .............CGGCACCTGCCGTTCGCTCCTGCTT.................................................................................................................................................. | 25 | 0 | 11 | 0.27 | 3 | 2 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................AGTGCCCCCACCCTTCCGCCGTCCCT.................................................................................... | 26 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ...........TGCGGCACCTGCCGTTCGCTCCT...................................................................................................................................................... | 23 | 0 | 11 | 0.18 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........GCTGCGGCACCTGCCGTTCGCTCC....................................................................................................................................................... | 24 | 0 | 11 | 0.18 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..CGCTGAGGCTGCGGCACCTGCCGTT............................................................................................................................................................. | 25 | 0 | 12 | 0.17 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................CGTCCCTCGTAACAGGGAGGCAGCT.................................................................. | 25 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...........TGCGGCACCTGCCGTTCGCTCCTGCT................................................................................................................................................... | 26 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........GGCTGCGGCACCTGCCGTTCGCTCCTC..................................................................................................................................................... | 27 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............GGCACCTGCCGTTCGCTCCTGCGT.................................................................................................................................................. | 24 | 1 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............CGCACCTGCCGGTCGTTCCTGCTT.................................................................................................................................................. | 24 | 3 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........GGCTGCGGCACCTGCCGTTCGCTCC....................................................................................................................................................... | 25 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............GGCACCTGCCGTTCGCTCCTGCTT.................................................................................................................................................. | 24 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......TGGCTGCGGCACCTGCCGTTCGCTCCT...................................................................................................................................................... | 27 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........AGCGGCACCTGCCGTTCGCTCCTGCTT.................................................................................................................................................. | 27 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............GCGGCACCTGCCGTTCGCTCCTG..................................................................................................................................................... | 23 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............GCGGCACCTGCCGTTCGCTCCTGCTTCT................................................................................................................................................ | 28 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................CACCTGCCGTTCGCTCCTGCTTCT................................................................................................................................................ | 24 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............GGCACCTGCCGTTCGCTCCTG..................................................................................................................................................... | 21 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........TGCGGCACCTGCCGTTCGCTCC....................................................................................................................................................... | 22 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........GGCTGCGGCCCCTGCCGTTCGCTCCT...................................................................................................................................................... | 26 | 1 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............GCACCTGCCGTTCGCTCCTGCTTCT................................................................................................................................................ | 25 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......GAGGCTGCGGCACCTGCCGTTCGCT......................................................................................................................................................... | 25 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............GGCACCTGCCGTTCGCTCCTGCTTCT................................................................................................................................................ | 26 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........GGCTGCGGCACCTGGCGTTCGCTCCT...................................................................................................................................................... | 26 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| CCCGCTGAGGCTGCGGCACCTGCCGTT............................................................................................................................................................. | 27 | 0 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........GGCTGCGGCACCTGCCGTTCG........................................................................................................................................................... | 21 | 0 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................GGAGGCAGCTACTGGGGCGGGGCCT................................................... | 25 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................CCTCGTAACAGGGAGGCAGCTACT............................................................... | 24 | 0 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................CTGTCTAGGGGCAGTGCCCCCACCCT................................................................................................ | 26 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................CCTGCCGTTCGCTCCTGCTTC................................................................................................................................................. | 21 | 0 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................GCCGTTCGCTCCTGCTTCT................................................................................................................................................ | 19 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................GTCTAGGGGCAGTGCCCCCACCCT................................................................................................ | 24 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................CCCGCCCTTCCGCCGTCCCT.................................................................................... | 20 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................CCCTTCCGCCGTCCCTCGTAACAGG........................................................................... | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................CGTGCCCCCACCCTTCCGCCGTCCCT.................................................................................... | 26 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................TCCGTCCCTCGTAACAGGGAGGCAGCT.................................................................. | 27 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................CCCCCACCCTTCCGCCGTCCCTC................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................CGCCGTCCCTCGTAACAGGGAGGCTGCT.................................................................. | 28 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................GGCAGTGCCCCCACCCTTCCGCCGTCCCT.................................................................................... | 29 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................CGCCGTCCCTCGTAACAGGGAGGCAGCT.................................................................. | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................TGCCCCCACCCTTCCGCCGTCCCT.................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................CCCCCACCCTTCCGCCGTCCCT.................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................TCCCGTCCCTCGTAACAGGGAGGCAGCT.................................................................. | 28 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_5676:2428-2611 + | dmo_383 | GGGCGACTCCGACGCCGTGGACGGCAAGC---GAGGAC-----------------------GAAGAAGCT-------------------CCGGCGGTAGAAGTGGACGACAGATCCCCGTCACGGGGGTGGGAAGGCGGCAGGGAGCATTGTCCCTCCGTCGATGACCCCGCCCCGGAGACCTGGCGCGG-----GATGGAGCGGAGGAAGGGC------GC-AGTGATGTGTCGAGAAGT |
| droVir3 | scaffold_13045:1167224-1167409 + | GGC-AACCCCAA--CCGCGGCCGGCGAGATGGA-CGAGGAAGAGTCCGCACCGTTCGGAACGGAGGAGCACACACCACCAGCGCAGGACGTGGAGGCGGTAGGAGAAGAAA--------------------------------------TAT--CCCCGTCGGC----------CGGAGACGCGGAGTCGCCACCTACGAAGGTGCCGAACGGCATCCGCGCAAACGACGGG-GAAGCAGA | |
| droAna3 | scaffold_13079:193747-193786 - | AGC-GAC----------------------------GAC-----------------------GAAGGAGCA-------------------GCGGCGGCAGCAGAAGACGACA---------------------------------------------------------------------------------------------------------------------------------- | |
| droFic1 | scf7180000453649:3241-3274 - | CGGCGGCTTCGTCGGAGAGGCCGGAGAGC---GAGGA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droTak1 | scf7180000414092:9572-9630 - | GGA-G--------------------------------------------------------------------------------------------------------------------------GGGAGGAGACGGCACAGAGAACTGTACCGCCAGCAACAACGGAGCCCCGGACATC----------------------------------------------------------- | |
| droEre2 | scaffold_4929:21632115-21632149 + | ACACGCCACCACCGCCGACGCGGGGAAGG---AAGGAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 10:11 PM