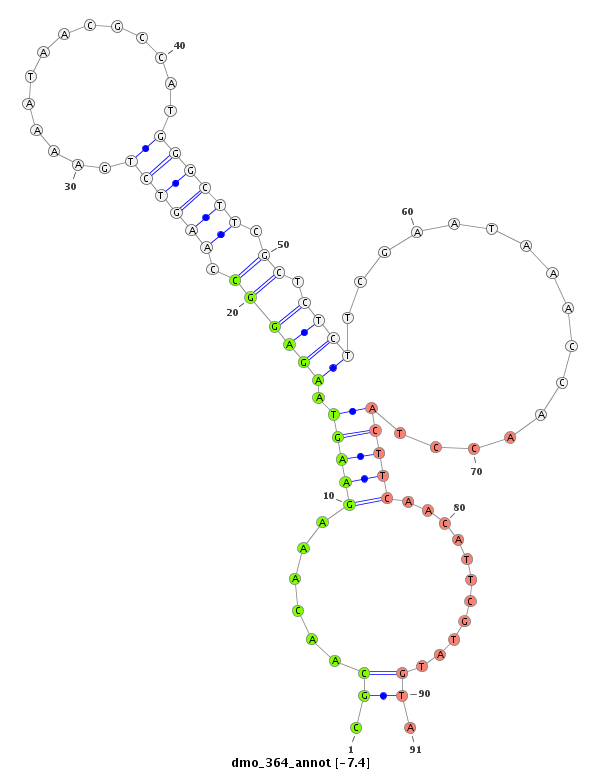
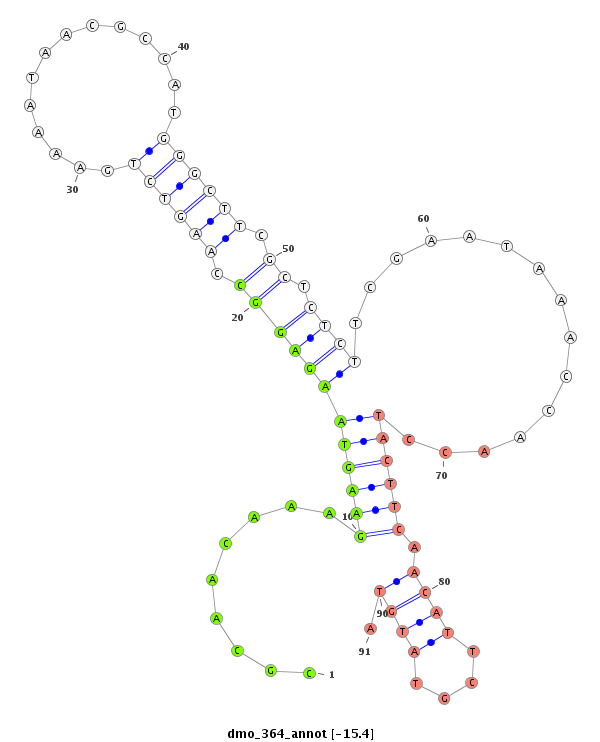
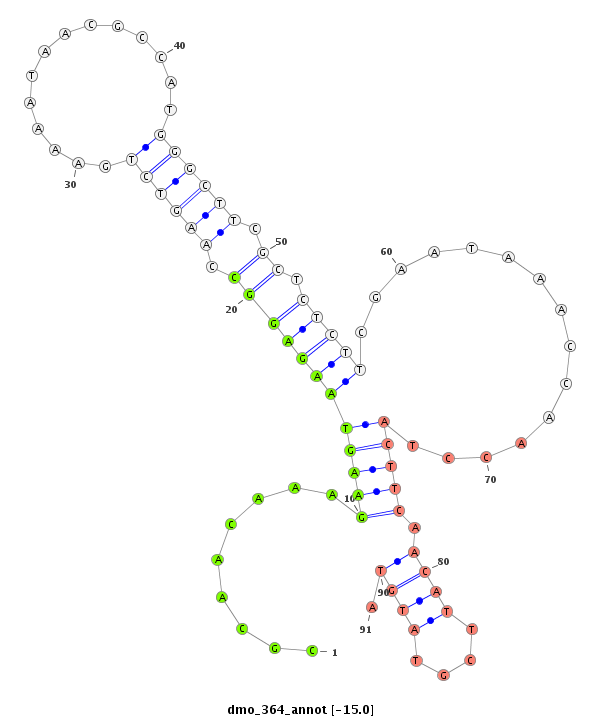
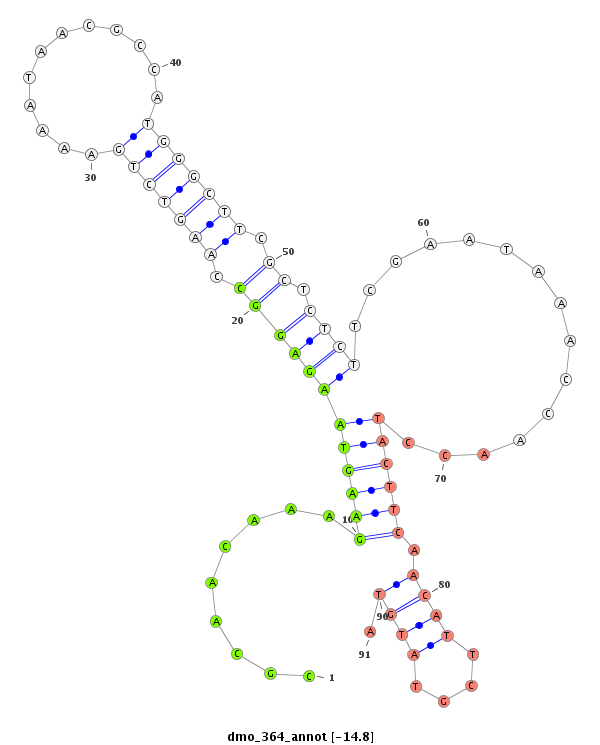
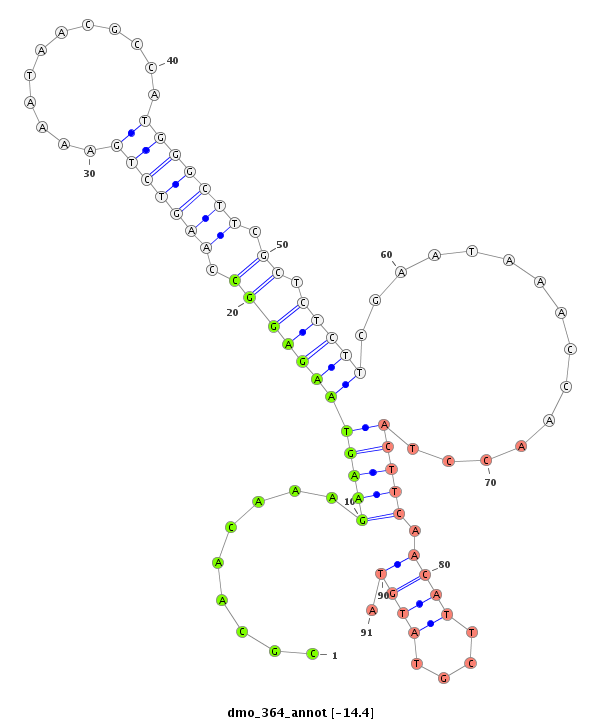
ID:dmo_364 |
Coordinate:scaffold_4314:2277-2427 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -15.4 | -15.0 | -14.8 | -14.4 |
 |
 |
 |
 |
exon [dmoj_GLEANR_13911:2]; CDS [Dmoj\GI13978-cds]; intron [Dmoj\GI13978-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## NNNNNNNNNNNNNNNNNNCTGTTTAATATATTGTGTTAAAAACCAAAAAGACAAAAATAAGTGTGTTCTGCCCGCTGGTACTGGAAGTGCCGCTGCTGCGGCACCCGCAACAAAGAAGTAAGAGGCCAAGTCTGAAAATAACGCCATGGGCTTCGCTCTCTTCGAATAAACCAACCTACTTCAACATTCGTATGTACTAAGCTTACGAAAACAACAACAGCAGCGACGGCAAGAACAACAGCAACCACGAA *********************************************************************************************************.((......(((((.((((((.((((((..............)))))).)).))))................)))))...........)).******************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V110 male body |
V056 head |
V049 head |
|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................CAAACCTACTTGAACATGCG............................................................. | 20 | 3 | 20 | 1.20 | 24 | 6 | 17 | 0 | 1 |
| ..........................................................................................................................................................................CAAACCTACTTGAACATGC.............................................................. | 19 | 3 | 20 | 0.90 | 18 | 18 | 0 | 0 | 0 |
| ............................................................................................................................................................................AACCTACTTGAACATGCG............................................................. | 18 | 2 | 20 | 0.15 | 3 | 2 | 0 | 1 | 0 |
| .........................................................................................................CGCAACAAAGAAGTAAGAGGC............................................................................................................................. | 21 | 0 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................CAAACCTACTTGAACATGCGT............................................................ | 21 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................ACCTACTTCAACATTCGTATGTA....................................................... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................GGCTTCGCTCTCTTCGAATA................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
NNNNNNNNNNNNNNNNNNGACAAATTATATAACACAATTTTTGGTTTTTCTGTTTTTATTCACACAAGACGGGCGACCATGACCTTCACGGCGACGACGCCGTGGGCGTTGTTTCTTCATTCTCCGGTTCAGACTTTTATTGCGGTACCCGAAGCGAGAGAAGCTTATTTGGTTGGATGAAGTTGTAAGCATACATGATTCGAATGCTTTTGTTGTTGTCGTCGCTGCCGTTCTTGTTGTCGTTGGTGCTT
*******************************************************.((......(((((.((((((.((((((..............)))))).)).))))................)))))...........)).********************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V110 male body |
V049 head |
V056 head |
|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................GTTTGGATGAACTTGTACG.............................................................. | 19 | 3 | 20 | 0.35 | 7 | 0 | 6 | 0 | 1 |
| ..........................................................................................................................................ATTGCGGTACCCGAAGCGAGAGAAGCT...................................................................................... | 27 | 0 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 |
| ...........................................................................................................................................TTGCGGTACCCGAAGCGAGAGAAGCTT..................................................................................... | 27 | 0 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 |
| .............................................................................................................................................GCGGTACCCGAAGCGAGAGAAGCTT..................................................................................... | 25 | 0 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 |
| ..........................................................................................................................................ATTGCGGTACCCGAAGCGAGAGAAGCTT..................................................................................... | 28 | 0 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 |
| ...........................................................TCACACAAGACGGGCGACCATGACCT...................................................................................................................................................................... | 26 | 0 | 10 | 0.30 | 3 | 3 | 0 | 0 | 0 |
| ......................................................................................................TGGGCGTTGTTTCTTCATTCTCCGGTT.......................................................................................................................... | 27 | 0 | 19 | 0.21 | 4 | 4 | 0 | 0 | 0 |
| ......................................................................................................TGGGCGTTGTTTCTTCATTCTCCGGT........................................................................................................................... | 26 | 0 | 19 | 0.16 | 3 | 3 | 0 | 0 | 0 |
| ..............................................................................................CGACGCCGTGGGCGTTGTTTCTTCATTCT................................................................................................................................ | 29 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 |
| .............................................................................................................................................GCGGTACCCGAAGCGAGAGAAGCTTAT................................................................................... | 27 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 |
| ..........................................................................................................................................................................GTTTGGATGAACTTGTACGC............................................................. | 20 | 3 | 20 | 0.15 | 3 | 1 | 0 | 2 | 0 |
| .............................................................................................................................................GCGGTACCCGAAGCGAGAGAAGCTTATT.................................................................................. | 28 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 |
| .............................................................................................................................................GCGGTACCCGAAGCGAGAGAAGCT...................................................................................... | 24 | 0 | 20 | 0.15 | 3 | 2 | 1 | 0 | 0 |
| .............................................................................................................................................GCGGTACCCGAAGCGAGAGAAGCTTATTT................................................................................. | 29 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 |
| .......................................................................................................GGGCGTTGTTTCTTCATTCTCCGGT........................................................................................................................... | 25 | 0 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 |
| ..................................................................................................................................................ACCCGAAGCGAGAGAAGCTTATTTGGTT............................................................................. | 28 | 0 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 |
| .......................................................................................................GGGCGTTGTTTCTTCATTCTCCGGTT.......................................................................................................................... | 26 | 0 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 |
| ............................................................................................................................................................................TTGGATGAACTTGTACGC............................................................. | 18 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| .........................................................ATTCACACAAGACGGGCGACCATGAC........................................................................................................................................................................ | 26 | 0 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................ACGACGCCGTGGGCGTTGTTTCTTCAT................................................................................................................................... | 27 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ..............................................................................................................................................CGGTACCCGAAGCGAGAGAAGCTT..................................................................................... | 24 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ............................................................................................................................................TGCGGTACCCGAAGCGAGAGAAGCTTAT................................................................................... | 28 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ........................................................TATTCACACAAGACGGGCGACCATGACCT...................................................................................................................................................................... | 29 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................GTACCCGAAGCGAGAGAAGCTTATTT................................................................................. | 26 | 0 | 20 | 0.10 | 2 | 1 | 1 | 0 | 0 |
| ..................................................................................................................................................ACCCGAAGCGAGAGAAGCTTATTTGGT.............................................................................. | 27 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| .................................................................................................CGCCGTGGGCGTTGTTTCTTCATTCT................................................................................................................................ | 26 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ..............................................................................................................................................CGGTACCCGAAGCGAGAGAAGCTTATTT................................................................................. | 28 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| .....................................................................................................................................................CGAAGCGAGAGAAGCTTATTTGGTT............................................................................. | 25 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................TTGCGGTACCCGAAGCGAGAGAAGCTTAT................................................................................... | 29 | 0 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................CCTGGGCGTTGTTTCTTCATTCTCCGGTT.......................................................................................................................... | 29 | 1 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................GTGGGCGTTGTTTCTTCATTCTCCGGTT.......................................................................................................................... | 28 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................TTTGGTTGGATGAAGTTGTAAGCAT........................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................CTTGGTTGGATGAAGTTGTAAGCATA.......................................................... | 26 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................TTGCGGTACCCGAAGCGAGAGAAGCT...................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................ACGCCGTGGGCGTTGTTTCTTCATTCT................................................................................................................................ | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................ATTGCGGTACCCGAAGCGAGAGAAGC....................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................TTCGGTACCCGAAGCGAGAGAAGCT...................................................................................... | 25 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................CCCGAAGCGAGAGAAGCTTATTTGGT.............................................................................. | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................TGCGGTACCCGAAGCGAGAGAAGCTTATTT................................................................................. | 30 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................CGACGCCGTGGGCGTTGTTTCTTCATT.................................................................................................................................. | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................CGACGCCGTGGGCGTTGTTTCTTCAT................................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................TGCGGTACCCGAAGCGAGAGAAGCTT..................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...................ACAAATTATATAACACAATTTTTGGTTTT........................................................................................................................................................................................................... | 29 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................TACCCGAAGCGAGAGAAGCTTATTT................................................................................. | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................GTACCCGAAGCGAGAGAAGCTT..................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................GACGCCGTGGGCGTTGTTTCTTCATT.................................................................................................................................. | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................CGACGACGCCGTGGGCGTTGTTTCTT...................................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................AGAGAAGCTTATTTGGTTGGATGAAGTT................................................................... | 28 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................................CACGGCGACGACGCCGTGGGCGTTGT........................................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................GTTCTTTTAGTCGTTGGTGCTT | 22 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................ACCCGAAGCGAGAGAAGCTTATT.................................................................................. | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................CGGTACCCGAAGCGAGAGAAGCTTATT.................................................................................. | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................GAAGCTTATTTGGTTGGATGAAGT.................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................ACCCGAAACGAGAGAAGCTTATTTGGT.............................................................................. | 27 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_4314:2227-2477 + | dmo_364 | NNNNNNNNNNNNNNNNNNCTGTTTAATATATTGTGTTAAAAACC----AAA--------AAGACAAAAA---------TAAGTGTGTTCTGCCCGCTGGTAC-TGGAAGTGCCGCTGCTGCG-------------------------------------------------------------------------------------------------GC---ACCCGCAACAAAGAAG-T-------AA------GA----------------------------GGCC--------AAGTCT--------GAAAATAACGCCATGGGCT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T------------------------------CGCTCTCT---------------------------------------------------------------------------------------T----CGAATAAACCAACCTACTTCAACATTCGTATGTACTAAGCTTA------------------------------------------------------------C------GAAAACAACAACAGCA--------------------GCGACGGCA------------------AGAACAACAGCA---AC---------------------------------------------------C------------ACGA-A |
| droVir3 | scaffold_12928:6658109-6658315 + | ------------------AC--------------------CAAA----AAG---------------AAGAAACCAAAGAAACAGCGCGCCGTCACCATGGAT-----G--------------------------------------------------A--------------------------------------------------------TGCGGC---CGTTGCGGCA-----------------------------------------------------------------------------------------------------------------------------GCGGCAACAACT-------------------------------------------------------------------------------------------------GGCGGCAA---------T-----------------------TTGCAGCGCATGCAACAGATA---------------------------------ACCAAATCAGCATCG-------------------------------------TCC---AACATGATGTCACAGCGTTTGCGCAG-C------------------------------AACATGGATTA------------------------------------------------------------C------TACAACAATAGCAGCA--------------------GCAGCAGCA------------------ACAACAACAGCA---AC---------------------------------------------------A------------ACAA-C | |
| droGri2 | scaffold_14753:2405-2587 - | ------------------AA--------------------AAAA----CAA---------------GAA---------------GGC------------------------------------------------------------------------------------------------AAAAACAA--CAGTAA---------------CAGTAAC---ATCAGCAGCA-----------------------------------------------------------------------------------------------------------------------------ACAGCAACAGCAGCAGCAGCAGCAAACAACAAGCTAAAA-ATGCCGTCAAATGCCA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAACACGTTTAAATGT------------------------------------------------------------------------------------CAAAATGTAACTCCGCAGAC------GGCAACAACAGCAACA--------------------GCAGTAGCA------------------GCAGCAAGAGTA---AC---------------------------------------------------A------------ACAG-C | |
| droWil2 | scf2_1100000004511:6264732-6264944 - | ------------------AA--------------------AACA----ACA--------------------------------------------------------------------AC---------A-------------------------------------------------------------------------------------------------A-----------------------------C--------------AACGATAAGCGCAG----------------------------CT------------GCA---------ACATCTACAACTGTTCCCTCAACCGGA-----------------------------------------------------------------------------------------------------------GCAGCAGCAGCAGCAGCTGG--AACAGCAACAACGACCAGCAGCAAGAA-ACTTACCGAGCATGATATTCAAGAGCGTTTGCTATCCTTCCATGCAGCCAACATTTCGTATTTGCAATCTCAGAATAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGGCA------------------AGTTCCTCCACT---AC---------------------------------------------------T------------ACATCT | |
| dp5 | XL_group1a:5701896-5702108 + | ------------------TC--------------------TGCC----AGC---------------T---------------------------------CG-AGGAAGAGGCGCGGCAACG-------------------------------------------------------------------------------------------------GC---AACGGCA---------TGGAGAACGAC------AG----------------------------GGAC--------AAGTCGGTTGAGCCCAAAGTAGAGCCAC--------------------------------------------------------------------------------------------------------------------------------------------------------------C-----------------------GGCCAGGA------------------------------CGCGCAGC---------------------------------------------------------------------------------------T----CCAGGCAGCAGACCAAGA----------------------------CC------CTCGACGTCCCATTGGGGGAAGAGCCATTACAGCGGGA----------------------GCCGCAGCAGCA--------------------GCCACAGGC------CTTGCCCACG--CCAACGCCAACGGCGAC-----------------------------------------------------------------CAC-T | |
| droPer2 | scaffold_12:202090-202251 - | ------------------TA--------------------CTGC----AAC-------------------------------------------------------------------------------A--------------------------------------CCAACGGAAGACAAA-----------------------------------AA---ATCAACA---------CCAA--------------A----------------------------AACCACGAAGGCTTCA--------------------------CCAACGATTTCACCCAAA---------AATTCAACTACAACGACA----------------------------------------------------------------ACA----------------------------------T---------CAA--------------------------------------------------------------------------------------------------------------------AGGTGAAA--------------------------------------------------------------------------TTAC------------------------------------------------------------CAAAAACTACAACAGAAAAGATA--------------------TTATCAACA------------------GCTGTGTCAACA---AC---------------------------------------------------C------------AAGA-C | |
| droAna3 | scaffold_13340:4279313-4279469 - | ------------------TG--------------------GCAG----C-------------------------------------------------------GGCAGTGTCGCTG---------------------------------------------------------------------------------G---------------CTGTGGC---AGTCACAACATTGTCGCC-------G-------GC----------------------------AGTC--------ATG---------------------------------------------------------------------------------ACCAGCTCGCCAATG-TCGCTGTCATGCATCCAACATACGGCAGTTG----------------------------------CCACAACAACAATA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAACAGCAACA--------------------GCAACAGCA------------------ATAACAACAACA---AC---------------------------------------------------A------------GCAG-T | |
| droBip1 | scf7180000396708:537482-537689 - | ------------------AG--------------------ATAC-------------------------------------TT---------------------------------------------------------TTGTCGCAGCTGCTGGCAATGGCAGC---ACA-----------------------------------------------GCTGTGACATCG---------TCAAGAATCACAAGCCC------------------------------------------TTGACC--------CGCA------------------------------------------------------ACTCCA--------------------------------------------------------------------------------------------------A---------G-----------------------TCAC------------------AGAGCCTTGTGGGCATCGGTCCATCCCAGTCAACCGTATCGCTGCCGTCGTACCCTCGCAGTGTAAGTGGCGAAAATAAGA--------------------------------------------------------------------------T-----------------------------------------------------------------------------CCCTCAACA--------------------CCAGCACCA------------------GAACAATCACCA---GA---------------------------------------------------T------------TCAA-A | |
| droKik1 | scf7180000302461:1897796-1897936 + | ------------------GC--------------------CAGA----TGG---------------T---------------------------------TG-AGGAAGCGGCTCTGCAGCA-------------------------------------------------------------------------------------------------GC---ATCTCCAACAAATCAC-G-------G-------GC-----------------AG-CGAAGCGT-------------------------------TGGCGCCAT----------------------------------------------TGTTGCTTCAGTTAGTGGGCCAAACTGTG----------CC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAATTGGCA--------------------GCAGAAACA------------------GCAACAGCAGCC---AC---------------------------------------------------A------------GCAA-C | |
| droFic1 | scf7180000454050:589706-589938 - | ------------------GT--------------------TCAAAATCAACGCCTGACAATGACAAAAG---------CAGCGACGTGGCGCCCCAAAATAA--------------------------------------------------------C-----------------------------------------------------------------AGCAG-----------------------------T-------------------------------------GGCAGCA--------------------------GCAGCAGCAGCAACTGAA---------GGAGCAGCAACAACAGCA----------------------------------------------------------------ACAGCAGTGCCAGGAG-------------------------------CAAC-----------------------------------------------------------------------------------------------------------------------------ACCAATCAAAA-----------------------------------------------------------------------------------------------------------------------------C------AGCAGCAACATCAAGA--------------------GCAGCAGCA------------------ACTGCAACTGCA---ACGAAAGCAATAACAATCAATGGGCCCCAAACGACATGGCGACGTCAACGAGGC------------TGCA-A | |
| droEle1 | scf7180000491261:641712-641899 - | ------------------AG--------------------AAGC----CAG---------------AAG---------AAAAT---------------------------------------------------------ATGTCTTGGAAAATGGAAAAGCCCGAGCAGCG-----------------------------------------------GC---AGCAATAACATTGAGC-T-------AG------TGATGTAATACGGAGCAAAAG-TGAAT-------------------------------------------CG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAAT---T--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGCGTTGGAAAAAAGAAACAGATTATACAATTTGTGTTGCTG-------------------------C-TGAAGTGAACTAGGACAACAGCAACAGCA------------------AT------ACTT---TC---------------------------------------------------A------------GC---G | |
| droRho1 | scf7180000766547:551-705 + | ------------------GG--------------------AAGC----C--------------------------------------------------------------GCGCTGCTGCA-------------------------------------------------------------------------------------------------GC---AACTGCAACAAATCAC-G-------G-------GC-----------------AG-CGAAGCGT-------------------------------TGGCGCCAT----------------------------------------------TGTTGCAGGAGTTAATAGGCCAAACTGTG----------CCA----------------------------------------AATTGGCAGGCACAGCAGAAGCA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAACAGCAGCA--------------------GCAACAACA------------------GCAACAGCAGCA---AC---------------------------------------------------A------------ACCA-A | |
| droBia1 | scf7180000302428:6867707-6867887 + | ------------------GT--------------------TGCT-----GC---------------T---------------------------------GG-AAGAAG-AACTCAGCGGCA---GCAACAGCAACAACCAAGCCAGGAACA-------------------------------------------------------------------AC---AGCAA-----------------------------C--------------AACACCAACAGCAG----------------------------CA------------GCA---------ACACTCAC-------------------------------------------------------------------------AGCAGCAACAACATAAG-------------------------------CA---ACAGCAACAGCAACA-------------------------------------------------------------------------------------------------------------------------------------------------------CC----AAAGCC-----------------------------------AG------------------------------------------------------------C------AGCAACAACGACACTA--------------------GCAGCAACA------------------GCAACAACAGCA---AC---------------------------------------------------A------------TCAG-C | |
| droTak1 | scf7180000412451:10788-10933 + | ------------------GC--------------------CACA------------------------------------------------------------------------GATGCAGCAAC---A--------------------------------------ACA-----------------------------------------------AC---AGGA------------------CTTTCAAAACC------------------------------------------AAGTCC--------AGCA------------ACG---------ACAACAGCATCAAAACCAACAACCACAGCA-------------------------------------------------------------------G-------------------------CCATACC---------AACAACAGCA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------G------------------------------------------------------------C------CAAACCAACAACAGCA--------------------GCCAAACCA------------------ACAACAACAGCA---GT---------------------------------------------------A------------ATAC-C | |
| droEug1 | scf7180000409538:100087-100283 - | ------------------CA--------------------AACG----------------------------------TAGCGGTGCCTTGCCCGCCAGCAACAGGCAGCA----------------------------------------------------------GCA-----------------------------------------------GC---AGCAGCAGC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGTTGCACAACGGCAATGGCAGTCAGAAT------------CC--CT----------------------------------------------------------------------------TCATCAAGGAGAACTACTGGAACACGCCGCC----CACAAGAGCCAGTCTGCTGCTCCATTCGCCTACGGAGT---------------------------------------------------------------------------ACTCTGAGGAGCA--------------------GCAGCAGCA------------------GCAACAACAACA---AC---------------------------------------------------A------------GCAG-C | |
| dm3 | chr2R:7853155-7853348 + | ------------------AA--------------------AAGA----CAA---------------AGA---------------TAT------------------------------------------------------------------------------------------------GACAATGC--CACCAA---------------TGGCAAC---AGCCATGGCA-----------------------------------------------------------------------------------------------------------------------------ACGGCAACAACAGCAGCA--------------------------------------------------------------------------------------------------A---------C-----------------------AACCAGCACAAAAGACAGCTAAC--GA----------------------------------------------------------------------------ACGCCAAGCAAGACAATGAGGACATGTTGAAAAG--------CAAAAATTATTTCAAT---AGTAAAAA----------------------------------------------------------------------------CAAAAGTATAGCAGCG--------------------GCAGCCGCA------------------AATGCAGCAGCA---GC---------------------------------------------------A------------GCAG-C | |
| droSim2 | 3r:21539608-21539803 + | ------------------AG--------------------AAGC----CAG---------------AAG---------AAAAT---------------------------------------------------------GTGTCTTGGAAAATGGAAAAGCACGA---GCA-----------------------------------------------GC---AGCTATAACATTGAGC-C-------AG------TGATGTAATACAGAGCAAAAG-TGAAGCGG-------------------------------------------------------------------------------------CG-------------------------------------------------------------------------------------------------------------------------------GCAAT---T--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGCATTGCAAAAAAGAAACAGATTATACAATTTGTGTTGCTG-------------------------C-TGCACTGAACTAGGACAACAGCAGCAACA------------------ACAGCAGCACTT---GG---------------------------------------------------A------------GCGG-C | |
| droSec2 | scaffold_0:6152490-6152695 + | ------------------AA--------------------AACC----AGC--------------------------------------------------------------------AC---------AACAACAAGCCCACGTGGT-------------------------------------ATTGCCGCAGCCGCAGCAATCTTCGTTGCTGCAGC---AATCGCAGCAGAGTC----------AG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAGCCCTTGGCTGTTATAGCAGCAG---------CAGCTGCT-----------------------G------------------------------CG--------------------------------------------------------------------------------------------CA----GCAGCC-----------------------------------CATGGCCACCTATTTTGT-------------------------------------T---------------------CGACAACC--------------------GCAGCAACAGCAACAACAGCAATCGCCAA------AGCC---GC---------------------------------------------------AAGCGCTTGTGGCACAA-C | |
| droYak3 | 3L:6409756-6409952 - | ------------------CA--------------------GCAA----C------------------------------ATGT---------------------------------------------------------GTGTTGTCGTCGTCGGCGACGACGC------------------AGCAGCTG--CAGCTG---------------CAGCAGC---ATCTGCAGCA-----------------------------------------------------------------------------------------------------------------------------ACAGCAACAGCAGCAGCA--------------------------------------------------------------------------------------------------G---------C-----------------------TTCC------------------------------------------------------------------------------------------------------------AAGATTATGGGAAGGCAGTGCC----GAAGAAA-----GCAGCTACTATATACCACTGAGTTCGGATAA------------------------------------------------------------C------GGCAGCGGGAGCAGTG--------------------AAAGCGCCG------------------AGAGCAGCAGCA---GC---------------------------------------------------A------------GTAG-C | |
| droEre2 | scaffold_4929:3967484-3967635 - | ------------------GC--------------------GGCG------------------------------------------------------------------------GCGGCGGCAGC---A--------------------------------------GCA-----------------------------------------------GC---AGCAGCGGCATTAACC-C----------------------------------------------------------AATGC-------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCAGCAA---------C-----------------------ATTCAGCA------------------------------CGCGCCAA---------------------------------------------------------------------------------------T----CTAACGAACCAACCCACC----------------------------AT------CGCCA-------------------------------------CTG-------------------------C-CAGCTCCAGCAACAACTACAACAACAACC---------------ACTTGAACAACAGCC---AC---------------------------------------------------A------------CCAC-C |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 10:08 PM