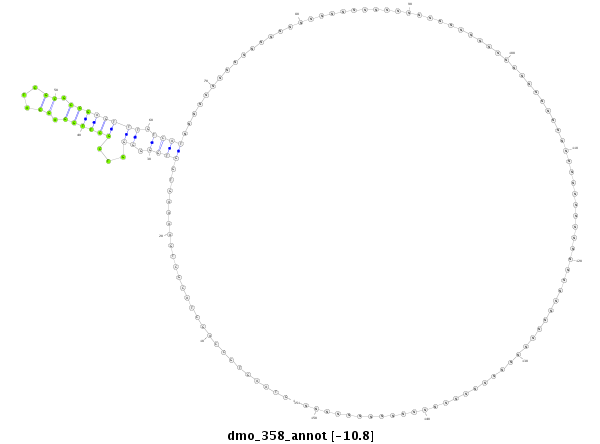
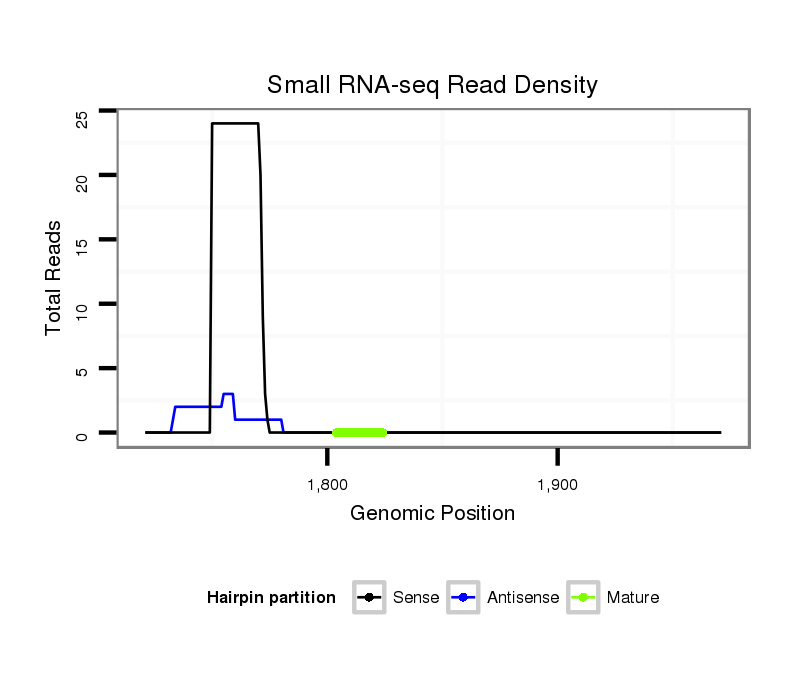
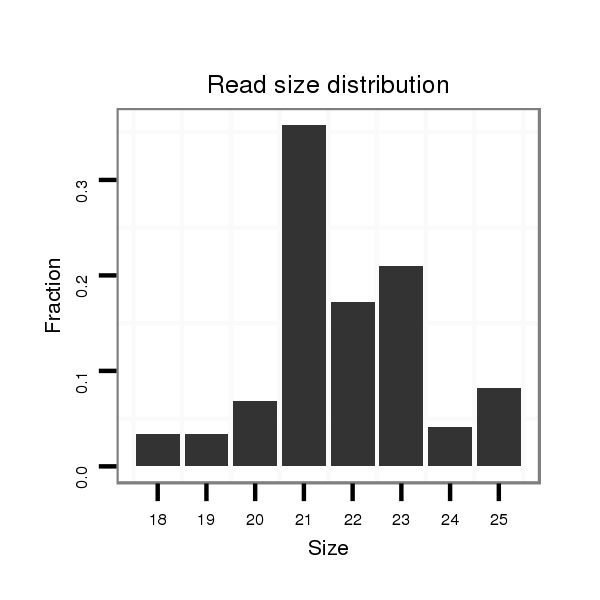
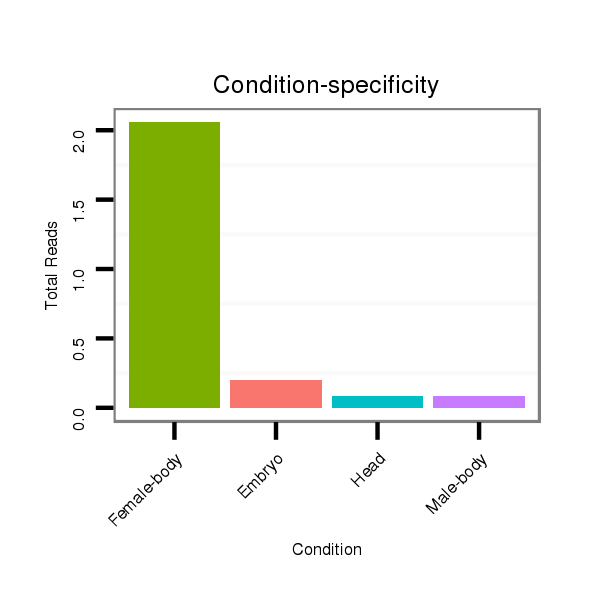
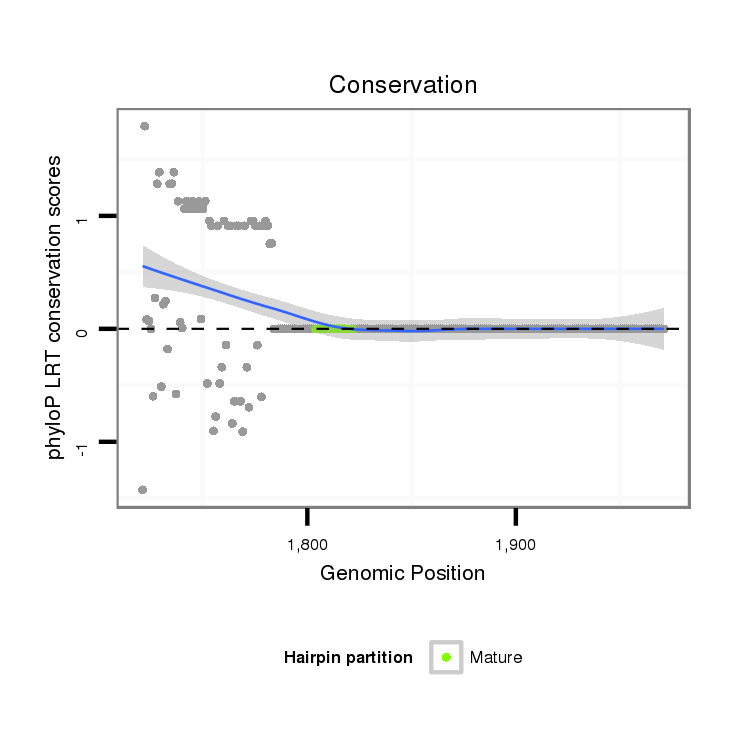
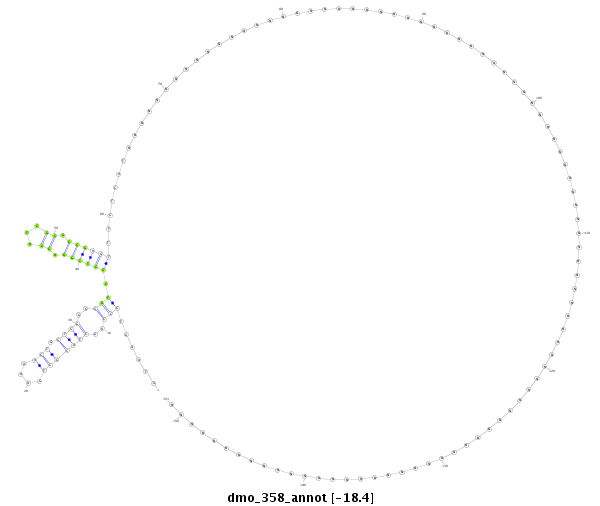
ID:dmo_358 |
Coordinate:scaffold_3911:1771-1921 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -18.4 | -18.4 | -17.7 |
 |
 |
 |
CDS [Dmoj\GI14267-cds]; exon [dmoj_GLEANR_14682:1]; intron [Dmoj\GI14267-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CGATAGTGATCTCCGATTCGGAGGAGGAAGAAACGCCAGAACCTCCGCGGGTAAGTGCCAGCTACGCTGAAAAGTGGTGGGGGGTCGCTTGGAGCATCGCGCCGAGTTTGTCATNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN **************************************************..........................((((.((...((((((.((...)).)))))))).)))).......................................................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V041 embryo |
V110 male body |
V049 head |
|---|---|---|---|---|---|---|---|---|---|---|
| .............................GAAACGCCAGAACCTCCGCGGG........................................................................................................................................................................................................ | 22 | 0 | 7 | 11.00 | 77 | 69 | 5 | 3 | 0 | 0 |
| .............................GAAACGCCAGAACCTCCGCGGGT....................................................................................................................................................................................................... | 23 | 0 | 7 | 6.00 | 42 | 36 | 5 | 0 | 1 | 0 |
| .............................GAAACGCCAGAACCTCCGCGG......................................................................................................................................................................................................... | 21 | 0 | 7 | 4.00 | 28 | 19 | 2 | 6 | 1 | 0 |
| .............................GAAACGCCAGAACCTCCGCGGGTA...................................................................................................................................................................................................... | 24 | 0 | 7 | 2.14 | 15 | 14 | 0 | 1 | 0 | 0 |
| .............................GAAACGCCAGAACCTCCGCGGGTAA..................................................................................................................................................................................................... | 25 | 0 | 7 | 1.57 | 11 | 11 | 0 | 0 | 0 | 0 |
| .............................GAAACGCCAGAACCTCCGCGGGC....................................................................................................................................................................................................... | 23 | 1 | 7 | 0.86 | 6 | 0 | 6 | 0 | 0 | 0 |
| ...................................................................................GTCGCTTGGAGCATCGCGCCG................................................................................................................................................... | 21 | 0 | 12 | 0.67 | 8 | 8 | 0 | 0 | 0 | 0 |
| .............................GAAACGCCAGAACCTCCGCGGGTAAG.................................................................................................................................................................................................... | 26 | 0 | 7 | 0.43 | 3 | 3 | 0 | 0 | 0 | 0 |
| ..............................AAACGCCAGAACCTCCGCGGG........................................................................................................................................................................................................ | 21 | 0 | 7 | 0.43 | 3 | 3 | 0 | 0 | 0 | 0 |
| ...................................................................................GTCGCTTGGAGCATCGCGCCGAG................................................................................................................................................. | 23 | 0 | 12 | 0.42 | 5 | 4 | 0 | 0 | 1 | 0 |
| ...................................................................................GTCGCTTGGAGCATCGCGCCGAGTTTC............................................................................................................................................. | 27 | 1 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...................................................................................GTCGCTTGGAGCATCGCGCCGA.................................................................................................................................................. | 22 | 0 | 12 | 0.33 | 4 | 3 | 0 | 0 | 0 | 1 |
| ......................................................................ATAGTGGTGGGGGGTCCC................................................................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................GAAACGCCAGAACCTCCGCG.......................................................................................................................................................................................................... | 20 | 0 | 7 | 0.29 | 2 | 1 | 1 | 0 | 0 | 0 |
| .............................GAAACGCCAGAACCTCCGCGGGTAAGT................................................................................................................................................................................................... | 27 | 0 | 7 | 0.29 | 2 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................................GTCGCTTGGAGCATCGCGCCGAGTT............................................................................................................................................... | 25 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................GTCGCTTGGAGCATCGCGCCGAGTTTA............................................................................................................................................. | 27 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................GAGCATCGCGCCGAGTTTGTC........................................................................................................................................... | 21 | 0 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................GTCGCTTGGAGCATCGCGCCGAGTC............................................................................................................................................... | 25 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................GGAACGCCAGAACCTCCGCGGGT....................................................................................................................................................................................................... | 23 | 1 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................GAAACGCCAGAACCTCCGCGGAC....................................................................................................................................................................................................... | 23 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................GAAACGCCAGAACCTCCGCGGGTATT.................................................................................................................................................................................................... | 26 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................AACGCCAGAACCTCCGCGGGTA...................................................................................................................................................................................................... | 22 | 0 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................GGAACGCCAGAACCTCCGCGGGC....................................................................................................................................................................................................... | 23 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................GAAACGCCAGCACCTCCGCGGG........................................................................................................................................................................................................ | 22 | 1 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................GAAACGCCAGAACCTCCGCGGGTAAGTGC................................................................................................................................................................................................. | 29 | 0 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................GGAACGCCAGAACCTCCGCGGG........................................................................................................................................................................................................ | 22 | 1 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................GAAACGCCAGAACCTCCGCGGGTAT..................................................................................................................................................................................................... | 25 | 1 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................GAAACGCCAGAACCTCCGCGGGAT...................................................................................................................................................................................................... | 24 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................AAACGCCAGAACCTCCGCGGGTA...................................................................................................................................................................................................... | 23 | 0 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................GAAACGCCAGAACCTCCGCGGGTAATT................................................................................................................................................................................................... | 27 | 1 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................GAAACGCCATAACCTCCGCGGGT....................................................................................................................................................................................................... | 23 | 1 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................GAAACGCCAGAACCTCCGCGTGT....................................................................................................................................................................................................... | 23 | 1 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................ACGCCAGAACCTCCGCGGG........................................................................................................................................................................................................ | 19 | 0 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................GAAACGCCGGAACCTCCGCGGGT....................................................................................................................................................................................................... | 23 | 1 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................TGGGGGGTCGCTTGGAGCATCGCG...................................................................................................................................................... | 24 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................GGGGGTCGCTTGGAGCATCGCGC..................................................................................................................................................... | 23 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................GTCGCTTGGAGCATCGCGCC.................................................................................................................................................... | 20 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................TCGCTTGGAGCATCGCGCCG................................................................................................................................................... | 20 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................TCGCTTGGAGCATCGCGCCGAG................................................................................................................................................. | 22 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................GTCGCTTGGAGCATCGCGCCGG.................................................................................................................................................. | 22 | 1 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................GTCGCTTGGAGCATCGCG...................................................................................................................................................... | 18 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................GTCGCTTGGAGCATCGCGC..................................................................................................................................................... | 19 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................GAAACTCCAGAACCTCCGCGGGT....................................................................................................................................................................................................... | 23 | 1 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................TCCGCCGGTAAGTGCCAGCTACGCT....................................................................................................................................................................................... | 25 | 1 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
|
GCTATCACTAGAGGCTAAGCCTCCTCCTTCTTTGCGGTCTTGGAGGCGCCCATTCACGGTCGATGCGACTTTTCACCACCCCCCAGCGAACCTCGTAGCGCGGCTCAAACAGTANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
**************************************************..........................((((.((...((((((.((...)).)))))))).)))).......................................................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V041 embryo |
V110 male body |
|---|---|---|---|---|---|---|---|---|---|
| ............GGCTAAGCCTCCTCCTTCTTTGCGGTC.................................................................................................................................................................................................................... | 27 | 0 | 5 | 1.80 | 9 | 9 | 0 | 0 | 0 |
| .............GCTAAGCCTCCTCCTTCTTTGCGGTC.................................................................................................................................................................................................................... | 26 | 0 | 5 | 1.40 | 7 | 7 | 0 | 0 | 0 |
| ..................................CGGTCTTGGAGGCGCCCATTCACGGT............................................................................................................................................................................................... | 26 | 0 | 7 | 1.00 | 7 | 5 | 0 | 1 | 1 |
| ..............CTAAGCCTCCTCCTTCTTTGCGGTC.................................................................................................................................................................................................................... | 25 | 0 | 5 | 0.80 | 4 | 0 | 3 | 0 | 1 |
| ...........AGGCTAAGCCTCCTCCTTCTTTGCGGTC.................................................................................................................................................................................................................... | 28 | 0 | 5 | 0.80 | 4 | 4 | 0 | 0 | 0 |
| ..........GAGGCTAAGCCTCCTCCTTCTTTGCGGTC.................................................................................................................................................................................................................... | 29 | 0 | 5 | 0.60 | 3 | 3 | 0 | 0 | 0 |
| ...................................GGTCTTGGAGGCGCCCATTCACGGT............................................................................................................................................................................................... | 25 | 0 | 14 | 0.43 | 6 | 5 | 0 | 1 | 0 |
| .......................................................................................GAACCTCGTAGCGCGGCTCAAACAGT.......................................................................................................................................... | 26 | 0 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 |
| .......................................TTGGAGGCGCCCATTCACGGTCGAT........................................................................................................................................................................................... | 25 | 0 | 15 | 0.40 | 6 | 3 | 3 | 0 | 0 |
| ...............TAAGCCTCCTCCTTCTTTGCGGTC.................................................................................................................................................................................................................... | 24 | 0 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 |
| .................................GCGGTCTTGGAGGCGCCCATTCACGGT............................................................................................................................................................................................... | 27 | 0 | 7 | 0.29 | 2 | 2 | 0 | 0 | 0 |
| ..............CTAAGCCTCCTCCTTCTTTGCGGGC.................................................................................................................................................................................................................... | 25 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| ............GGCTAAGCCTCCTCCTTCTTTGCGG...................................................................................................................................................................................................................... | 25 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................GAACCTCGTAGCGCGGCTCAAACAG........................................................................................................................................... | 25 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ...............TAAGCCTCCTCCTTCTTTGCGGGC.................................................................................................................................................................................................................... | 24 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| ...........CGGCTAAGCCTCCTCCTTCTTTGCGGTC.................................................................................................................................................................................................................... | 28 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 |
| ..........TTGGCTAAGCCTCCTCCTTCTTTGCGGTC.................................................................................................................................................................................................................... | 29 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .............GCTAAGCCTCCTCCTTCTTTGCGGT..................................................................................................................................................................................................................... | 25 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ................AAGCCTCCTCCTTCTTTGCGGTC.................................................................................................................................................................................................................... | 23 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .........................................GGAGGCGCCCATTCACGGTCGAT........................................................................................................................................................................................... | 23 | 0 | 15 | 0.20 | 3 | 3 | 0 | 0 | 0 |
| .............................................................................................CGTAGCGCGGCTCAAACAGTA......................................................................................................................................... | 21 | 0 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| .........AGAGGCTAAGCCTCCTCCTTCTTTGCGGT..................................................................................................................................................................................................................... | 29 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ...............TAAGCCTCCTCCTTCCTTGC........................................................................................................................................................................................................................ | 20 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ......................................CTTGGAGGCGCCCATTCACGGTCGAT........................................................................................................................................................................................... | 26 | 0 | 15 | 0.20 | 3 | 2 | 1 | 0 | 0 |
| ...............................................................................................TAGCGCGGCTCAAACAGTA......................................................................................................................................... | 19 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ............GGCTAAGCCTCCTCCTTCTTTGCGGT..................................................................................................................................................................................................................... | 26 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .............GCTAAGCCTCCTCCTTCTTT.......................................................................................................................................................................................................................... | 20 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ............GGCTAAGCCTCCTCCTTCTTC.......................................................................................................................................................................................................................... | 21 | 1 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 |
| ..............CTAAGCCTCCTCCTTCTTT.......................................................................................................................................................................................................................... | 19 | 0 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 |
| ..............CTAAGCCTCCTCATTCTTTGGGGTC.................................................................................................................................................................................................................... | 25 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 |
| .................................GCGGTCTGGGAGGCGCCCATTCACGGT............................................................................................................................................................................................... | 27 | 1 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 |
| ............................TCTTTGCGGTCTTGGAGGCGCCCAT...................................................................................................................................................................................................... | 25 | 0 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 |
| .................................GCGGTCTTGGAGGCGCCCATT..................................................................................................................................................................................................... | 21 | 0 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| ..................................CGTTCTTGGAGGCGCCCATTCACGGT............................................................................................................................................................................................... | 26 | 1 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 |
| ......................................TTTGGAGGCGCCCATTCACGGTCGAT........................................................................................................................................................................................... | 26 | 1 | 15 | 0.13 | 2 | 2 | 0 | 0 | 0 |
| .......................................TTGGAGGCGCCCATTCACGGT............................................................................................................................................................................................... | 21 | 0 | 15 | 0.13 | 2 | 1 | 1 | 0 | 0 |
| ......................................TTTGGAGGCGCCCATTCACGGT............................................................................................................................................................................................... | 22 | 1 | 15 | 0.13 | 2 | 2 | 0 | 0 | 0 |
| ...........AGGCTAAGCCTCCTCCGTCTTTGCGGTC.................................................................................................................................................................................................................... | 28 | 1 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 |
| ............GGCTAAGCCTCCTCCGTCTTTGCGGTC.................................................................................................................................................................................................................... | 27 | 1 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 |
| ..............CTAAGCCTCCTCCATCTTTGCGGTC.................................................................................................................................................................................................................... | 25 | 1 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 |
| ....................................GTCTTGGAGGCGCCCATTCACGGT............................................................................................................................................................................................... | 24 | 0 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| ...................................GGTCTTGGAGGCGCCCATTCATGTT............................................................................................................................................................................................... | 25 | 2 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 |
| ......................................GTTGGAGGCGCCCATTCACGGT............................................................................................................................................................................................... | 22 | 1 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| .........................................GGAGGCGCCCATTCACGGTCGA............................................................................................................................................................................................ | 22 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| .......................................TTGGAGGCGCCCATTCACGGTCGTT........................................................................................................................................................................................... | 25 | 1 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 |
| ........................................TGGAGGCGCCCATTCACGGTCGAT........................................................................................................................................................................................... | 24 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| .......................................TTGGAGGCGCCCATTCACGGTCGATGC......................................................................................................................................................................................... | 27 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| .........................................GGAGGCGCCCATTCACGGTCGATGC......................................................................................................................................................................................... | 25 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| .......................................TTGGAGGCGCCCATTCACGGTC.............................................................................................................................................................................................. | 22 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| .......................................TTGGAGGCGCCCATTCACGGTCGA............................................................................................................................................................................................ | 24 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| ......................................CTTGGAGGCGCCCATTCACGG................................................................................................................................................................................................ | 21 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| .......................................TTGGAGGCGCCCATTCACGG................................................................................................................................................................................................ | 20 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| ........................................CGGAGGCGCCCATTCACGGTCGAT........................................................................................................................................................................................... | 24 | 1 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 |
| .........................................GGAGGCGCCCAATCACGGTCGAT........................................................................................................................................................................................... | 23 | 1 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| ........................................TGGAGGCGCCCATTCACGGTCGATGC......................................................................................................................................................................................... | 26 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| .......................................TTGGAGGCGCCCATTCACGGTCGATGCG........................................................................................................................................................................................ | 28 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| .........................................TGAGGCGCCCATTCACGGTCGAT........................................................................................................................................................................................... | 23 | 1 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ............................................GGCGCCCATTCACGGTCGATT.......................................................................................................................................................................................... | 21 | 1 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ......................................TTTTGAGGCGCCCATTCACGGTCGAT........................................................................................................................................................................................... | 26 | 2 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ...........................................AGGCGCCCATTCACGGTCGATGCGAC...................................................................................................................................................................................... | 26 | 0 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 |
| ................................................................................CTACAGCGAACCTCGTATC........................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_3911:1721-1971 + | dmo_358 | CGATAGTGATCTCCGATTCGGAGGAGGAAGAAACGCCAGAACCTC-------CGCGGGTAAGTGCCAGCTACGCTGAAAAGTGGTGGGGGGTCGCTTGGAGCATCGCGCCGAGTTTGTCATNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN |
| droVir3 | scaffold_12928:7456959-7457026 - | AGGTGATGAACTTCGATTCCGAGGAGGAGGACACCGCCAAGCCCATGTCCTACGATGAAAAGCGACAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15110:5773339-5773369 + | AGACGGAGAGCGCCGAGTTCGAGGAGGAGGA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004511:1204227-1204232 - | CGATAG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droPer2 | scaffold_8:1965830-1965833 + | TGAT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_13117:2158871-2158940 - | AGGTGATGAACTTCGACTCCGAGGAGGAGGACACGGCCAAGCCGATGTCGTACGACGAGAAGCGCCAGCT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000415878:580788-580803 + | TGAGGCTGAATTTCGA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4690:9823759-9823760 + | CG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 10:03 PM