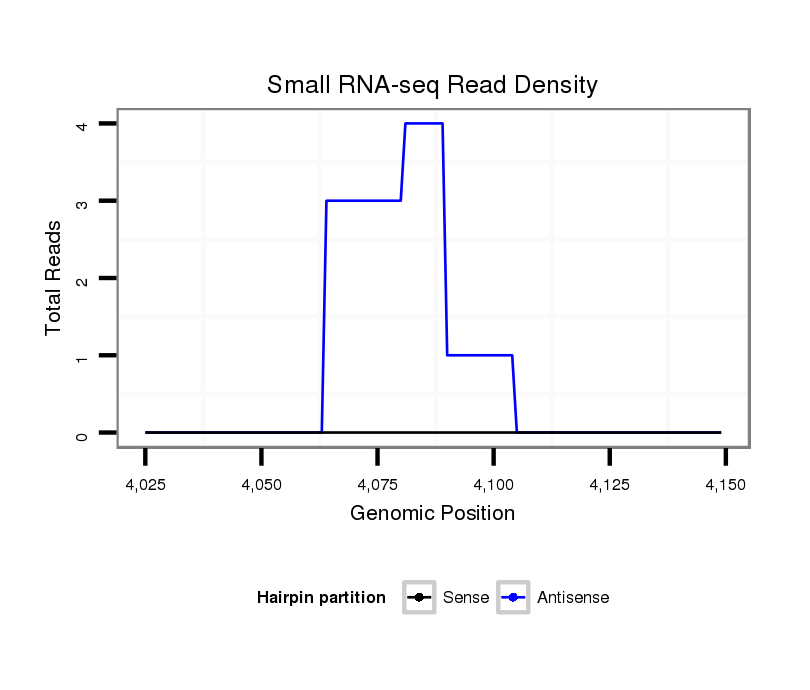
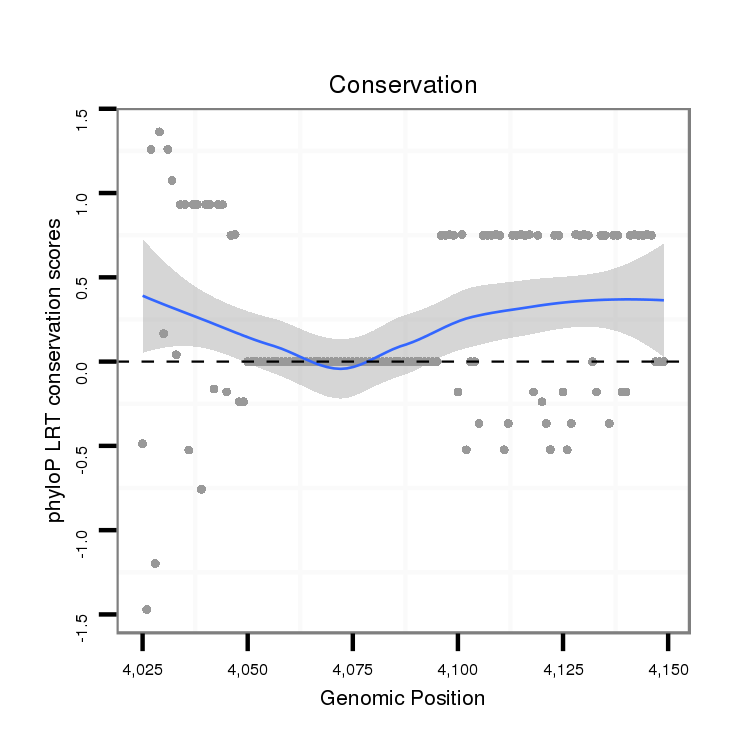
ID:dmo_355 |
Coordinate:scaffold_1834:4075-4099 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
intron [Dmoj\GI21867-in]; CDS [Dmoj\GI21867-cds]; exon [dmoj_GLEANR_6974:2]; exon [dmoj_GLEANR_6974:1]; CDS [Dmoj\GI21867-cds]
No Repeatable elements found
| ##################################################-------------------------################################################## TCCGTCCCAGGTGGAGGGCCACATTATCTCAACAGATGAGGAGTTAGAGTGTTGGGCCAGCCCGATACCGTGCAGATGAACGCGAGGAGGAGAGCAACGCGGAAGAGGTCCCCGACCAGCTCCCG **************************************************((((((....)))))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V110 male body |
V041 embryo |
|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................GCAACGCGGAAGAGGTCCCCGACCAAC..... | 27 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ..........................................GTTAGAGTGTTGGGCCAGCCCGA............................................................ | 23 | 0 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 |
| .............................................................................................GCAACGCGGAAGAGGTCCCCG........... | 21 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| .........................................AGTTAGAGTGTTGGGCCAGCC............................................................... | 21 | 0 | 4 | 0.50 | 2 | 0 | 2 | 0 | 0 |
| .............................................................................................GCAACGCGGAAGAGGTCCCCGACCAGC..... | 27 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| .............................................................................................GCCACGCGGAAGAGGTCCCCGACCAGC..... | 27 | 1 | 4 | 0.50 | 2 | 0 | 2 | 0 | 0 |
| ...........................................TTAGAGTGTTGGGCCAGCC............................................................... | 19 | 0 | 4 | 0.50 | 2 | 0 | 1 | 0 | 1 |
| ..........................................CTTAGAGTGTTGGGCCAGCCCG............................................................. | 22 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| .................................................TGTTGGGCCAGCCCGATACCGTG..................................................... | 23 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ..........................................GTTAGAGTGTTGGGCCAGCCCGATACC........................................................ | 27 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ...........................................TTAGAGTGTTGGGCCAGCCCGA............................................................ | 22 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ...........................................TTAGAGTGTTGGGCCAGCCCGAT........................................................... | 23 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ..........................................GTTAGAGTGTTGGGCCAGCCC.............................................................. | 21 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ..........................................GTTAGAGTGTTGGGCCAGCCCGTA........................................................... | 24 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................CGCGGAAGAGGTCCCCGACCAGCTC... | 25 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ..........................................GTTAGAGTGTTGGGCCAGCCCGAT........................................................... | 24 | 0 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| .......................................GGAGTTAGAGTGTTGGGCCA.................................................................. | 20 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ......................................AGGTGTTAGAGTGTTGGGCC................................................................... | 20 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| ...........................................TTAGAGTGTTGGGCCAGCCCG............................................................. | 21 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ......................................AGGAGTTAGAGTGTTGGGCT................................................................... | 20 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................................................CGGAAGAGGTCCCCGACCAGC..... | 21 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................TGAGGTCCCCGACCAGCTCCC. | 21 | 1 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 |
|
AGGCAGGGTCCACCTCCCGGTGTAATAGAGTTGTCTACTCCTCAATCTCACAACCCGGTCGGGCTATGGCACGTCTACTTGCGCTCCTCCTCTCGTTGCGCCTTCTCCAGGGGCTGGTCGAGGGC
**************************************************((((((....)))))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V110 male body |
|---|---|---|---|---|---|---|---|---|
| .......................................CCTCAATCTCACAACCCGGTCGGGCT............................................................ | 26 | 0 | 4 | 3.25 | 13 | 9 | 4 | 0 |
| ........................................................GGTCGGGCTATGGCACGTCTACTT............................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 |
| ......GTTCCACCTCACCGTGTAATAGAGT.............................................................................................. | 25 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| ........................................CTCAATCTCACAACCCGGTCGGGCT............................................................ | 25 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| .......................................CCTCAATCTCACAACCCGGTCGGGCTAT.......................................................... | 28 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| .....................................CTCCTCAATCTCACAACCCGG................................................................... | 21 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| .......................................CCTCAATCTCACAACCCGGTCGGGC............................................................. | 25 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| .................................................................................CGCTCCTCCTCTCGTTGCGCCT...................... | 22 | 0 | 10 | 0.10 | 1 | 0 | 0 | 1 |
| .....................................................................................CCTCCTCTCGATGCGCCTTCTCCAG............... | 25 | 1 | 12 | 0.08 | 1 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_1834:4025-4149 + | dmo_355 | TCCGTCCCAG------GTGGAGGGCCACATTATCTCAACAGATGAGGAGTTAGAGTGTTGGGCCAGCCCGATACCGTGCAGATGAACGCGAGGAGGAGAGCAACGCGGAAGAGGTCCCCGACCAGCTCCCG |
| droVir3 | scaffold_12730:229235-229243 - | TGCATCCCA-------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_572:10339-10345 + | TCCGTCC---------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000777572:10956-11034 - | GGCCTACCCGAACCATGAGGCGGGCCGCAAA----------------------------------------------GCAGGTC--AGCGAGCCGGAGAACTCGGCACCAGAG-CCCACGGTCAGCTC--- | |
| droEre2 | scaffold_4679:32127-32146 + | CCCATCCCCG------GTGGAGGACC--------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 10:01 PM