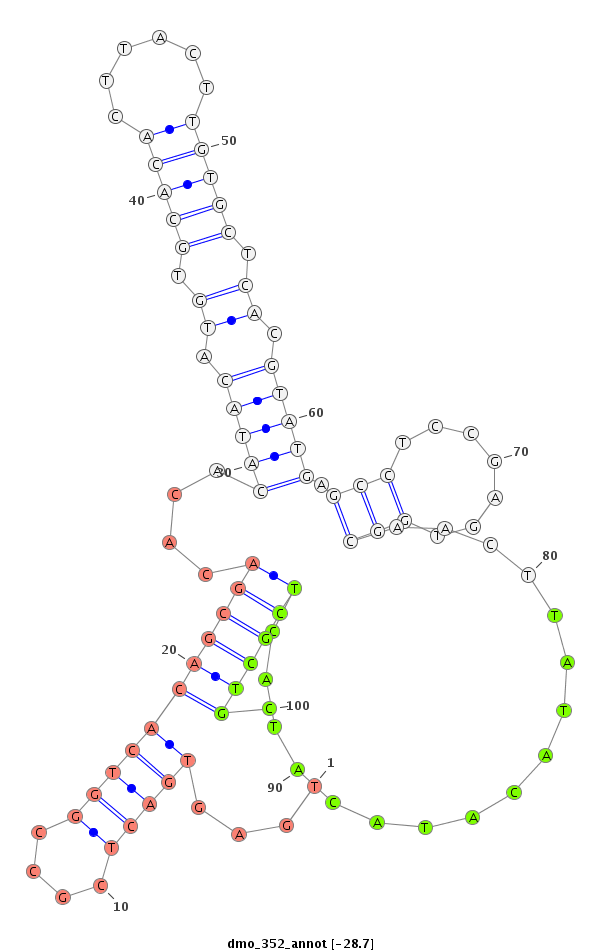
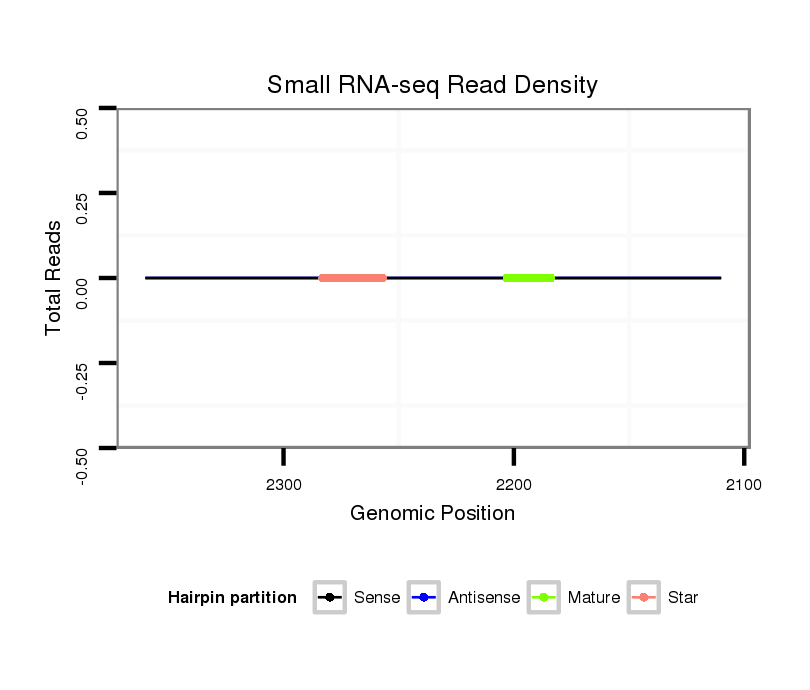
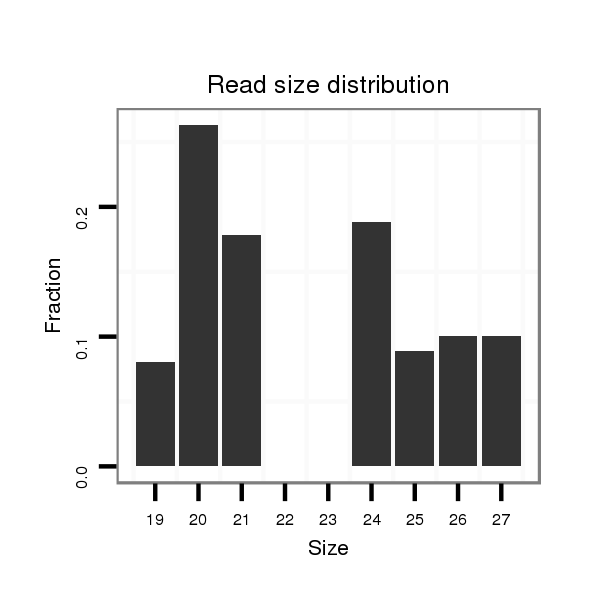
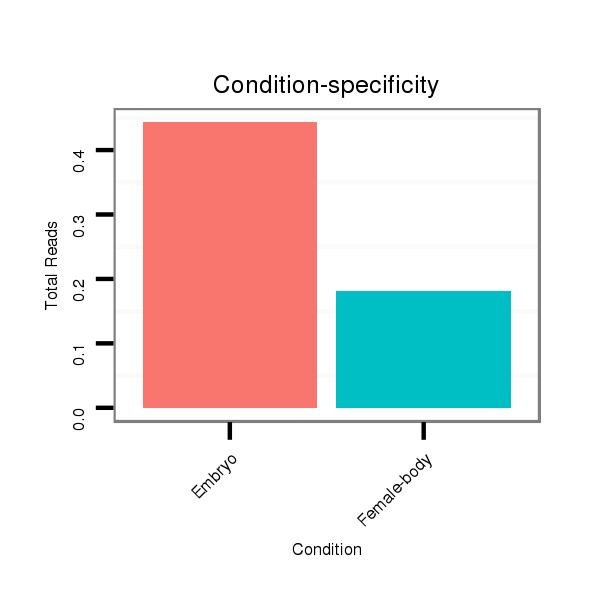
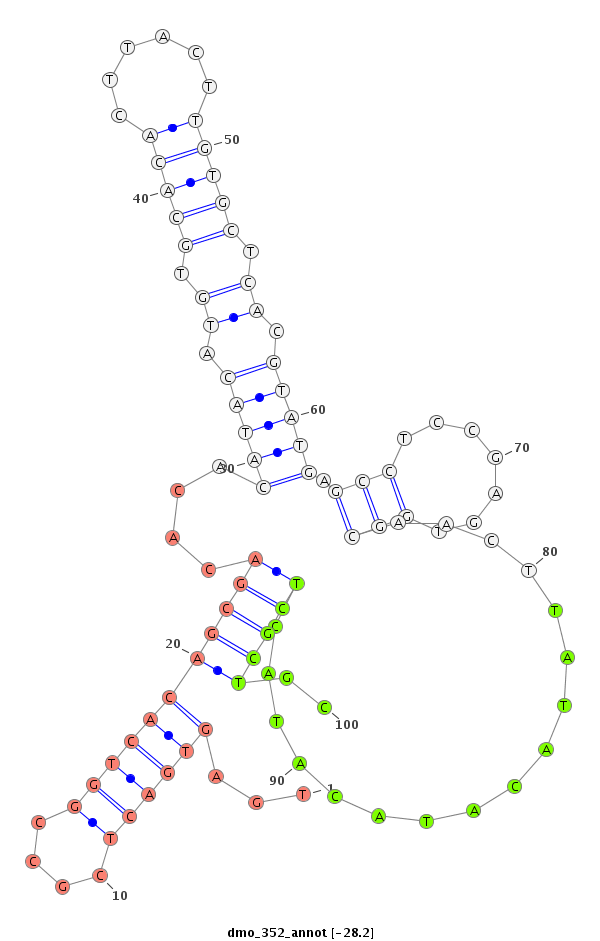
ID:dmo_352 |
Coordinate:scaffold_2747:2160-2310 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
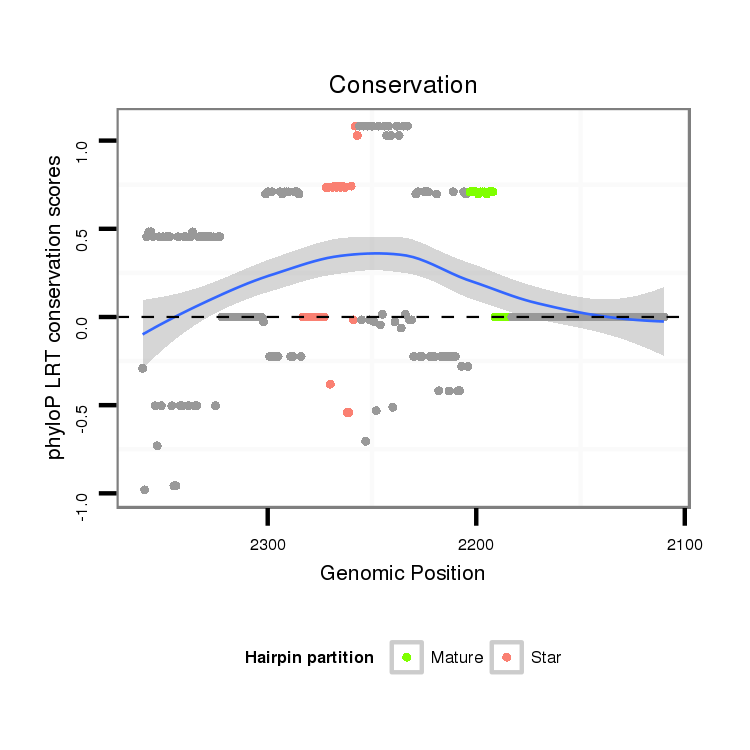
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
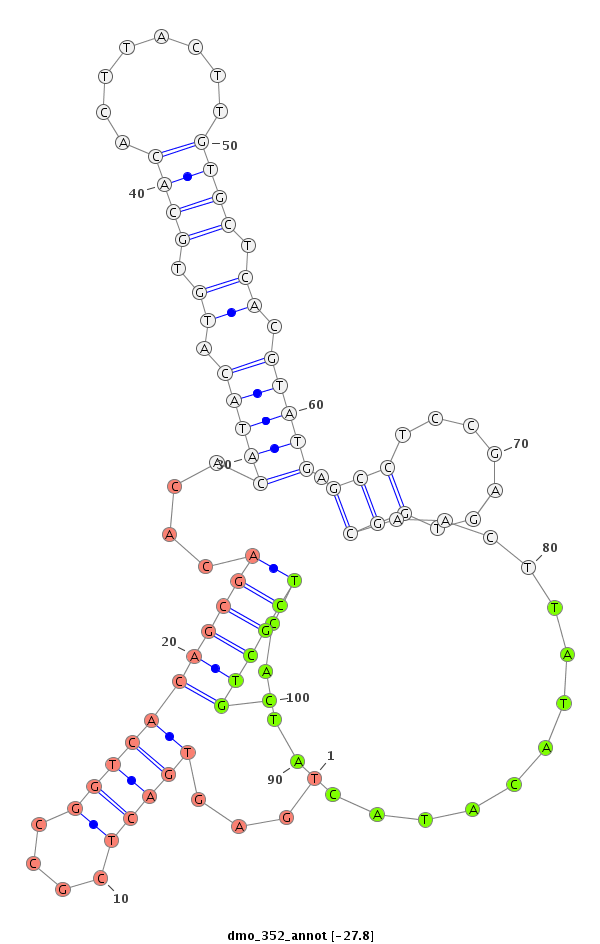
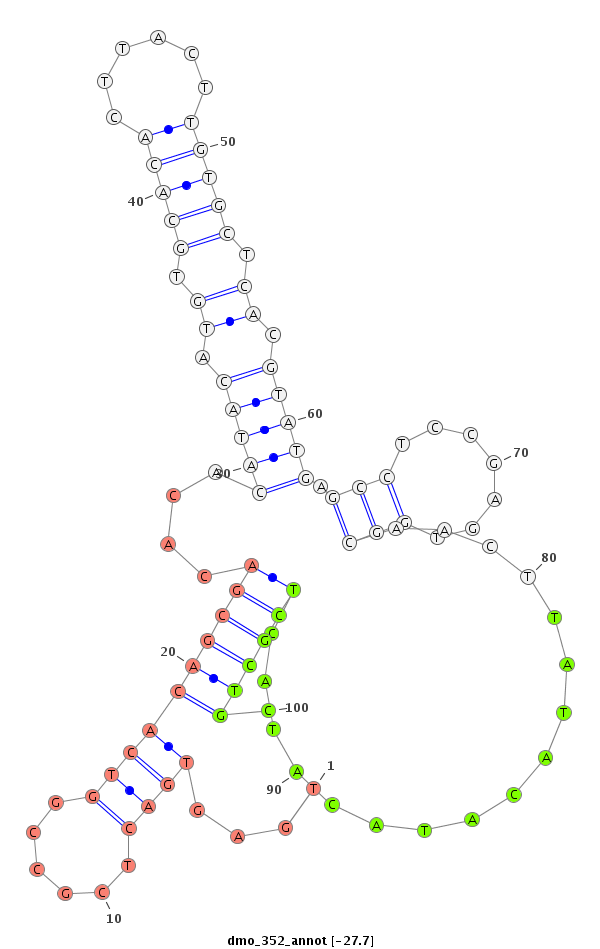
| -28.2 | -27.8 | -27.7 |
 |
 |
 |
CDS [Dmoj\GI23907-cds]; exon [dmoj_GLEANR_91:1]; intron [Dmoj\GI23907-in]
| Name | Class | Family | Strand |
| Galileo_DM | DNA | P | - |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CTCTTCGGGAGGCGCGGCGTCGAGTATGGGCGGCCGCGAGTTACCCGCCGGTCACTATACACACACACATACGTACATGAGTGACTCGCCGGTCACAGCGACACACATACATGTGCACACTTACTTGTGCTCACGTATGAGCCTCCGAGTGGCAACTTATACATACATACTCGCTGCTGCTTGTCGTCAGCATGAAAACGGAATAAAGTTCAGTAGATTGTTCATTTGACTCGAAACAAGACGTCTTTTAT *****************************************************************************....(((((....)))))((((((....(((((.((.(((((......))))).)).))))).(((.......))).................)))))).************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M060 embryo |
M046 female body |
V041 embryo |
V110 male body |
|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................TATACATACATACTCGCTGC.......................................................................... | 20 | 0 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 |
| ..................................................................................GACTCGCCGGTTACAGCGACACA.................................................................................................................................................. | 23 | 1 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................TGAGTGACTCGCCGGTCACAGCGACAC................................................................................................................................................... | 27 | 0 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| .........................................................................TACATGAGTGACTCGCCGGTCACAGC........................................................................................................................................................ | 26 | 0 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................TCACGTATGAGCCTCCGAGT..................................................................................................... | 20 | 0 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 |
| ..................................................................ACATACGTACATGAGTGACTCGCC................................................................................................................................................................. | 24 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................CAGCGACACACATACATGTGC....................................................................................................................................... | 21 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| .............................................................CACACACATACGTACATGAGTGAC...................................................................................................................................................................... | 24 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................ACTCGCTGCTGCTTGTCGTCAGCAT.......................................................... | 25 | 0 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| .......................................................AATACACACACACATACGTACATGAG.......................................................................................................................................................................... | 26 | 1 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .............................................................CACACACATACGTACATGAGT......................................................................................................................................................................... | 21 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................TATTCATACATACTCGCTGC.......................................................................... | 20 | 1 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ............................................................AGACACACATACGTACATGAGT......................................................................................................................................................................... | 22 | 1 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................TATACATACATACTCGCTGCAA........................................................................ | 22 | 2 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................TACATGTGCACACTTACTTGC........................................................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................ACACATACATGTGCACACT.................................................................................................................................. | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................TTTGACTCGAAACAAGACGTCTTTC.. | 25 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
|
GAGAAGCCCTCCGCGCCGCAGCTCATACCCGCCGGCGCTCAATGGGCGGCCAGTGATATGTGTGTGTGTATGCATGTACTCACTGAGCGGCCAGTGTCGCTGTGTGTATGTACACGTGTGAATGAACACGAGTGCATACTCGGAGGCTCACCGTTGAATATGTATGTATGAGCGACGACGAACAGCAGTCGTACTTTTGCCTTATTTCAAGTCATCTAACAAGTAAACTGAGCTTTGTTCTGCAGAAAATA
**************************************************************************....(((((....)))))((((((....(((((.((.(((((......))))).)).))))).(((.......))).................)))))).***************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V110 male body |
M060 embryo |
V041 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................CGAACAGCAGTCGTACTTTTGCCTT................................................ | 25 | 0 | 17 | 0.53 | 9 | 8 | 0 | 1 | 0 |
| ...................................................................................................................................................................................GAACAGCAGTCGTACTTTTGCCTTATTT............................................ | 28 | 0 | 16 | 0.31 | 5 | 5 | 0 | 0 | 0 |
| .................................................................................................................................................................................ACGAACAGCAGTCGTACTTTTGCCTT................................................ | 26 | 0 | 17 | 0.24 | 4 | 4 | 0 | 0 | 0 |
| ........................................AATGGGCGGCCAGTGATATGTGTGT.......................................................................................................................................................................................... | 25 | 0 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................GGCTCACCGTTGAATATGTATGTAT.................................................................................. | 25 | 0 | 16 | 0.19 | 3 | 0 | 3 | 0 | 0 |
| ..............................................................................................................................................GAGGCTCACCGTTGAATATGTATGTAT.................................................................................. | 27 | 0 | 16 | 0.19 | 3 | 0 | 2 | 1 | 0 |
| .............................................................................................................................................GGAGGCTCACCGTTGAATATGTATG..................................................................................... | 25 | 0 | 16 | 0.19 | 3 | 2 | 1 | 0 | 0 |
| ............................................................................................................................................CGGAGGCTCACCGTTGAATATGTATGT.................................................................................... | 27 | 0 | 16 | 0.19 | 3 | 1 | 2 | 0 | 0 |
| .....................................................................................................................................................................................ACAGCAGTCGTACTTTTGCCTTATTT............................................ | 26 | 0 | 17 | 0.18 | 3 | 2 | 0 | 1 | 0 |
| ..............................................................................................................................................GAGGCTCACCGTTGAATATGTATGT.................................................................................... | 25 | 0 | 16 | 0.13 | 2 | 1 | 1 | 0 | 0 |
| ...............................................................................................................................................AGGCTCACCGTTGAATATGTATG..................................................................................... | 23 | 0 | 16 | 0.13 | 2 | 2 | 0 | 0 | 0 |
| .............................................................................................................................................GGAGGCTCACCGTTGAATATGTATGT.................................................................................... | 26 | 0 | 16 | 0.13 | 2 | 0 | 2 | 0 | 0 |
| ......................................................................................................................................................................................CAGCAGTCGTACTTTTGCCTTATTT............................................ | 25 | 0 | 17 | 0.12 | 2 | 2 | 0 | 0 | 0 |
| ...................................................................................................................................................................................GAACAGCAGTCGTACTTTTGCCTT................................................ | 24 | 0 | 17 | 0.12 | 2 | 2 | 0 | 0 | 0 |
| .................................GGCGCTCAATGGGCGGTCAGTGAT.................................................................................................................................................................................................. | 24 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 |
| .........TCCGCGCCGCAGCTCATACCC............................................................................................................................................................................................................................. | 21 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 |
| ...............CCGCTGCTCCTACCCTCCGG........................................................................................................................................................................................................................ | 20 | 3 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 |
| ..................................................................................................................................................................................TGAACAGCAGTCGTACTTTTGCCTTAT.............................................. | 27 | 1 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................GGCTCACCGTTGAATATGTATGT.................................................................................... | 23 | 0 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................CGAACAGCAGTCGTACTTTTGCCTTATT............................................. | 28 | 0 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................CACCGTTGAATATGTATGTATGAGC.............................................................................. | 25 | 0 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................GGAGGCTCACCGTTGAATATGTATGTAT.................................................................................. | 28 | 0 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................CGGAGGCTCACCGTTGAATATGTAT...................................................................................... | 25 | 0 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................CTCGGAGGCTCACCGTTGAATAT.......................................................................................... | 23 | 0 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................CCGTTGAATATGTATGTATGAGCGA............................................................................ | 25 | 0 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................AGGCTCACCGTTGAATATATATGTAT.................................................................................. | 26 | 1 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................GGAGGCTCACCGTTGAATATTTATGT.................................................................................... | 26 | 1 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................AGGCTCACCGTTGAATATGTATGTAT.................................................................................. | 26 | 0 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................CGGAGGCTCACCGTTGAATATGTATGTAT.................................................................................. | 29 | 0 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................CCGTTGAATATGTAAGTATGAGCGA............................................................................ | 25 | 1 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................GGAGGCTCACCGTTGAATATGTAGG..................................................................................... | 25 | 1 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................GAGGCTCACCGTTGAGTATGTATGTAT.................................................................................. | 27 | 1 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................TCACCGTTGAATATGTATGTATGAGC.............................................................................. | 26 | 0 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................CGGAGGCTCACCGTTGAATATGTATG..................................................................................... | 26 | 0 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................CAGCAGTCGTACTGTTGCCTTATTT............................................ | 25 | 1 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................GAACAGCAGTCGTACTTTTGCCT................................................. | 23 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................GACGAACAGCAGTCGTACTTTTGCCTT................................................ | 27 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................GCAGTCGTACTTTTGCCTTATT............................................. | 22 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................CAGCAGTCGTACTTTTGCCTTATT............................................. | 24 | 0 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................................ACAGCAGTCGTACTTTTGCCTTATT............................................. | 25 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................TGAATATGTATGTATGAGCGACGAC........................................................................ | 25 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................CAGCGGTCGTACTTTTGCCTTATTT............................................ | 25 | 1 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................GAACAGCAGTCGTACTTTTGCCTTA............................................... | 25 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................GCAGTCGTACTTTTGCCTTATTT............................................ | 23 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................AGCAGTCGTACTTTTGCCTTATTT............................................ | 24 | 0 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................GGCCAGTGTCGCTGTGTGTATTT............................................................................................................................................ | 23 | 1 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| .......................................................................................CGGCCAGTGTCGCTGTGTGT................................................................................................................................................ | 20 | 0 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................AGTGCCTTCTCGGAGCCTC...................................................................................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................ATGTATGAGCGACGACGAACAGCAGT.............................................................. | 26 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................GTATGAGCGACGACGAACAGCAGT.............................................................. | 24 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................ACGTGTGAATGAACACGAGTGCAT.................................................................................................................. | 24 | 0 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ..................................................CAGTGATATGTGTGTGTGTATGCAT................................................................................................................................................................................ | 25 | 0 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................TGTATGAGCGACGACGAACAGCAGT.............................................................. | 25 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................CGGTCAGTGTCGGTGTGTGTA............................................................................................................................................... | 21 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................AGTAAACTGAGAAGTGTTCTG......... | 21 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................GTAAACTGAGCTTTGTTCTGCAGA..... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_2747:2110-2360 - | dmo_352 | CTCT------------TCGGGAGGCGCGGCGTCGAGTA-TGGGCGGCCGCGAGTTACCCGCCGGTCACTATACACACACACATACGTACATGAGTGACTCGCCGGTCACAGCGACACACATACATGTGCACACTTACTTGTGCTCACGTATGAGCCTCCGAGTGGCAACTTATACATACATACTCGCTGCTGCTTGTCGTCAGCATGAAAACGGAATAAAGTTCAGTAGATTGTTCATTTGACTCGAAACAAGACGTCTTTTAT |
| droVir3 | scaffold_12736:510027-510077 + | CGCTAAAACTCCGCCATCATGGGGCGTCCCACCGGGTGGCGGGCGGCCACG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000001063:3300-3388 + | AC---------------------------------------------------------------------ACATATGTACATATATACG------------------------TACATACATATTTACACACATACATGTATCCATATATAGACACTTACATATATACATATACATACATA---------------------------------------------------------------------------------- | |
| droKik1 | scf7180000302698:1431372-1431416 + | TT---------------------------------------------------------------------G-----------------------------CCTGTCACAGGCACACACAGACACATATACACGCACATATGC------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
Generated: 05/17/2015 at 10:05 PM