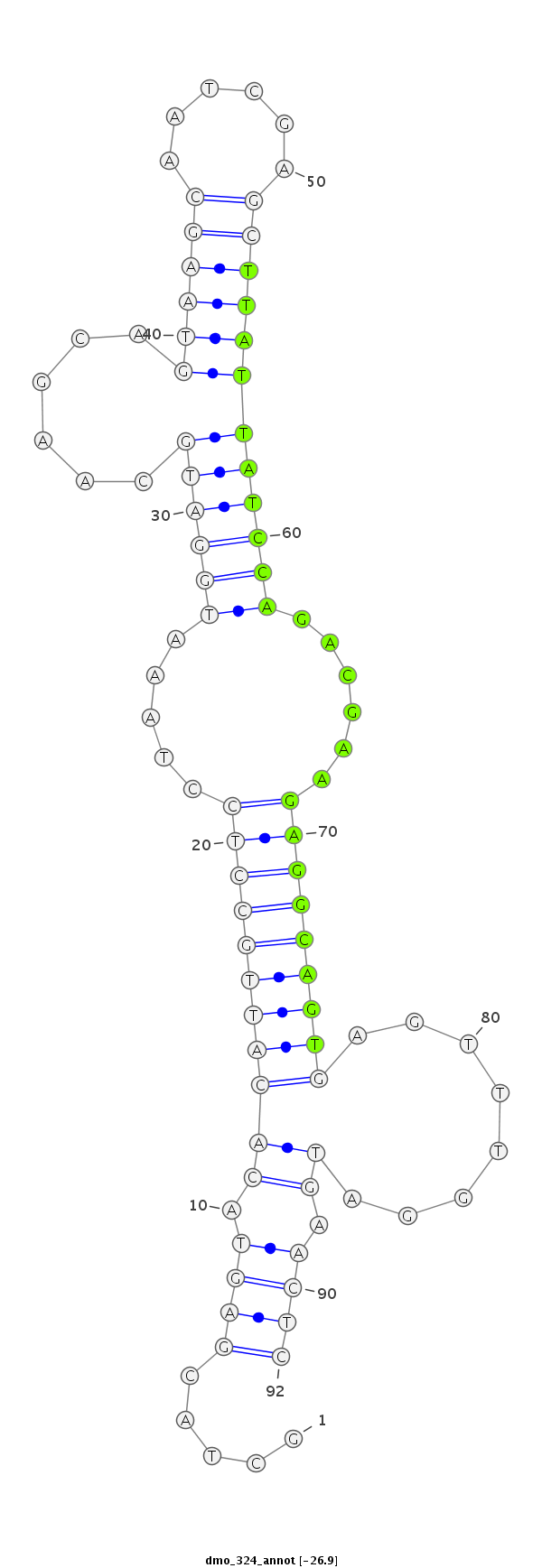
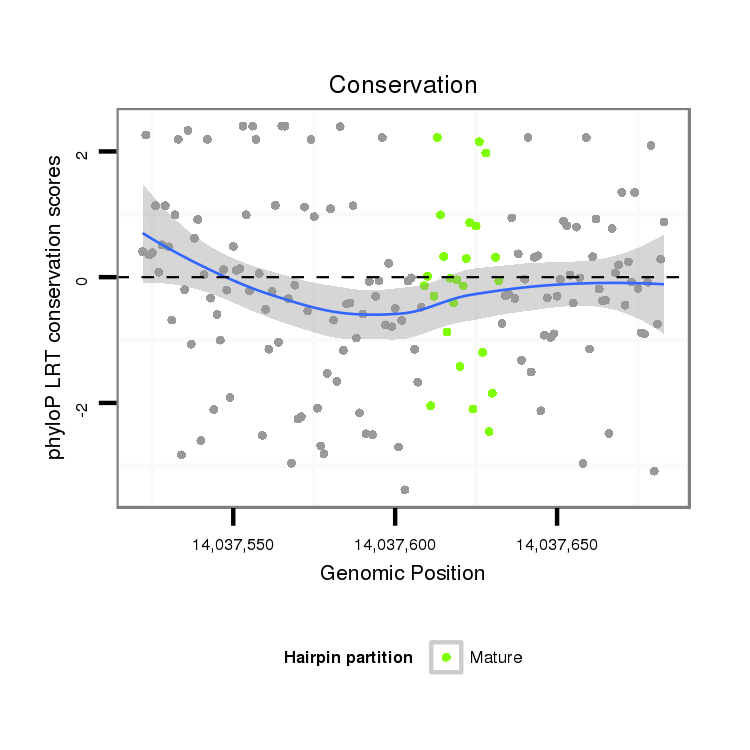
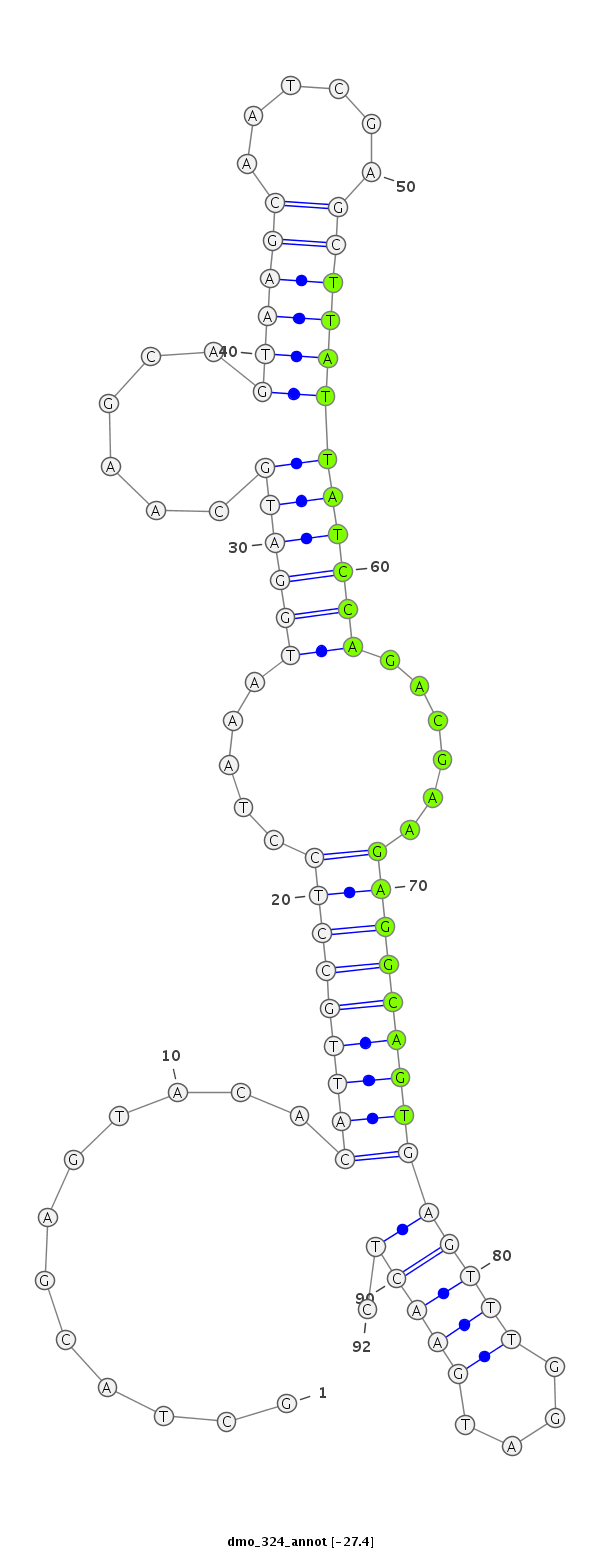
ID:dmo_324 |
Coordinate:scaffold_6500:14037572-14037633 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
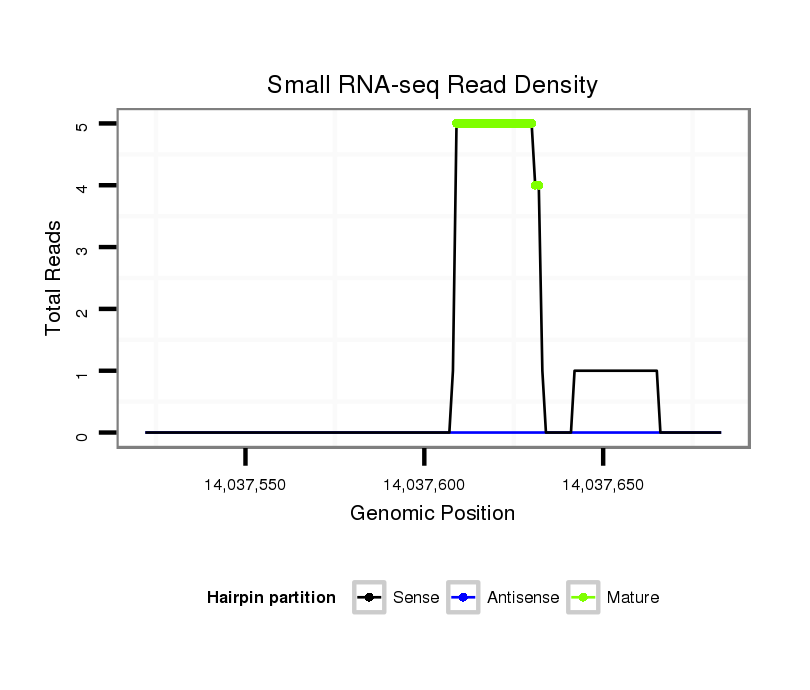
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -27.4 | -27.0 | -26.8 |
 |
 |
 |
Antisense to intron [Dmoj\GI11243-in]
No Repeatable elements found
| mature | star |
|
TTTTAGATACGGACTAAGTCGTAACAACAAATGCTGCTACGAGTACACATTGCCTCCTAAATGGATGCAAGCAGTAAGCAATCGAGCTTATTATCCAGACGAAGAGGCAGTGAGTTTGGATGAACTCATAAATGAGCTGCAGGAGTAATCAAATTTAACATT
***********************************.....((((.(((((((((((.....((((((......((((((......))))))))))))......)))))))))........)).))))*********************************** |
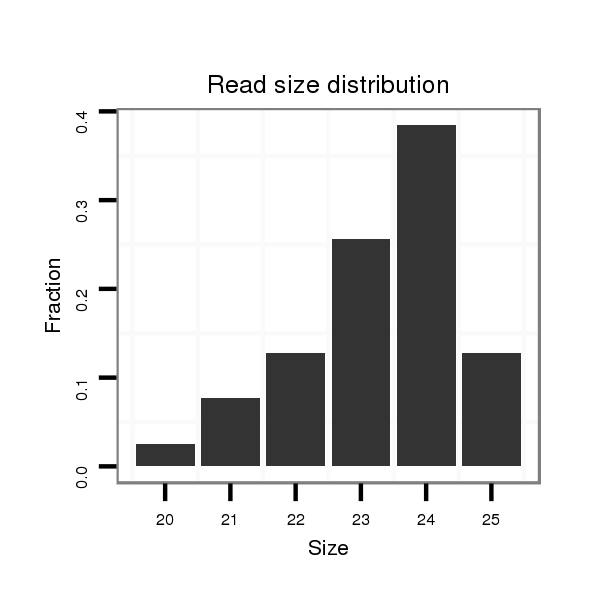
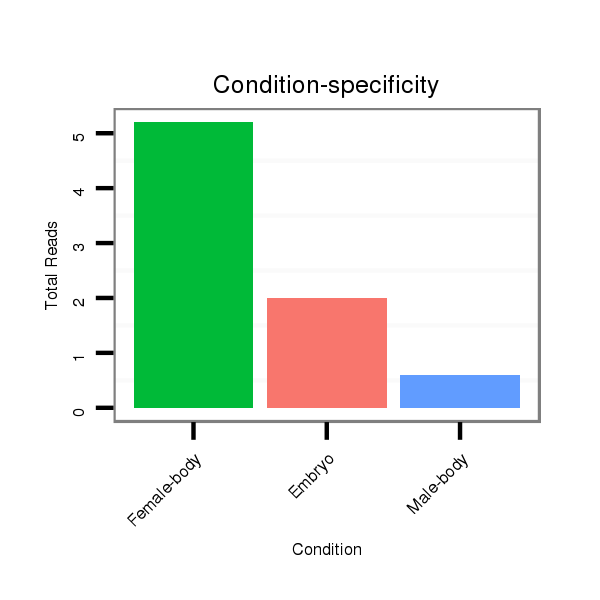
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V110 male body |
V041 embryo |
|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................TTATTATCCAGACGAAGAGGCAGT................................................... | 24 | 0 | 5 | 3.00 | 15 | 11 | 1 | 3 | 0 |
| .......................................................................................TTATTATCCAGACGAAGAGGCAGTG.................................................. | 25 | 0 | 5 | 1.00 | 5 | 4 | 1 | 0 | 0 |
| ......................................................................................CTTATTATCCAGACGAAGAGGCA..................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ........................................................................................................................TGAACTCATAAATGAGCTGCAGGA.................. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................TTATTATCCAGACGAAGAGGCA..................................................... | 22 | 0 | 5 | 0.80 | 4 | 3 | 0 | 0 | 1 |
| ........................................................................................TATTATCCAGACGAAGAGGCAGT................................................... | 23 | 0 | 5 | 0.60 | 3 | 3 | 0 | 0 | 0 |
| .......................................................................................TTATTATCCAGACGAAGAGGC...................................................... | 21 | 0 | 5 | 0.60 | 3 | 2 | 1 | 0 | 0 |
| .......................................................................................TTATTATCCAGACGAAGAGGCAGTGAG................................................ | 27 | 0 | 5 | 0.60 | 3 | 2 | 1 | 0 | 0 |
| .......................................................................................TGATTATCCAGACGAAGAGGCAGT................................................... | 24 | 1 | 5 | 0.40 | 2 | 0 | 2 | 0 | 0 |
| .......................................................................................TTATTATCCAGACGAAGAGGCAG.................................................... | 23 | 0 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 |
| .......................................................................................TTATTATCCAGACGAAGAGGCAGTGAGT............................................... | 28 | 0 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 |
| .......................................................................................TTATTATCCAGACGAAGAGGCAGTGA................................................. | 26 | 0 | 5 | 0.40 | 2 | 0 | 0 | 2 | 0 |
| ...........................................................................................TATCCAGACGAAGAGGCAGTGAG................................................ | 23 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................TGATTATCCAGACGAAGAGG....................................................... | 20 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| .......................................................................................TTATTATCCAGACGAAGAGG....................................................... | 20 | 0 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| .......................................................................................TGATTATCCAGACGAAGAGGC...................................................... | 21 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................ATCCAGACGAAGAGGCAGTGAGTTTGG........................................... | 27 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................TATTATCCAGACGAAGAGGCAG.................................................... | 22 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................TCCAGACGAAGAGGCAGTGAGTTTGG........................................... | 26 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................TTATTATCCAGACGAAGAGCCA..................................................... | 22 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................GTTATTATCCAGACGAAGAGGCAGT................................................... | 25 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................TTATTATCCAGACGCAGAGGC...................................................... | 21 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| .......................................................................................TTATTATCCAGACGAAAAGGCAGTG.................................................. | 25 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................TTATTATCCAGACGAAGAGGCAGTGAA................................................ | 27 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ......................AGCAACAAATGGTGATACGA........................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
AAAATCTATGCCTGATTCAGCATTGTTGTTTACGACGATGCTCATGTGTAACGGAGGATTTACCTACGTTCGTCATTCGTTAGCTCGAATAATAGGTCTGCTTCTCCGTCACTCAAACCTACTTGAGTATTTACTCGACGTCCTCATTAGTTTAAATTGTAA
***********************************.....((((.(((((((((((.....((((((......((((((......))))))))))))......)))))))))........)).))))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
|---|---|---|---|---|---|---|---|
| .............................................................................................AGGTCTGCTTCTCCGTCACTCAAACCT.......................................... | 27 | 0 | 5 | 0.80 | 4 | 3 | 1 |
| .................................................................................................CTGCTTCTCCGTCACTCAAACCTACT....................................... | 26 | 0 | 5 | 0.60 | 3 | 2 | 1 |
| ..............................................................................................GGTCTGCTTCTCCGTCACTCAAACCT.......................................... | 26 | 0 | 5 | 0.60 | 3 | 2 | 1 |
| ....................................................................................................CTTCTCCGTCACTCAAACCT.......................................... | 20 | 0 | 5 | 0.60 | 3 | 0 | 3 |
| .................................................................................................CTGCTTCTCCGTCACTCAAACCT.......................................... | 23 | 0 | 5 | 0.60 | 3 | 3 | 0 |
| ...............................ACGACGATGCTCATGTATAACGGAGGAT....................................................................................................... | 28 | 1 | 4 | 0.25 | 1 | 1 | 0 |
| .........................................TCATGTGTAACGGAGGATTTACGT................................................................................................. | 24 | 1 | 4 | 0.25 | 1 | 0 | 1 |
| ...............................ACGACGATGCTCATGTGTAACGGAGGAT....................................................................................................... | 28 | 0 | 4 | 0.25 | 1 | 1 | 0 |
| ..................................ACGATGCTCATGTGTAACGGAGGATT...................................................................................................... | 26 | 0 | 4 | 0.25 | 1 | 0 | 1 |
| ...................................CGATGCTCATGTGTAACGGAGGAT....................................................................................................... | 24 | 0 | 4 | 0.25 | 1 | 1 | 0 |
| ........................................................................................ATAATAGGTCTGCTTCTCCGTCACT................................................. | 25 | 0 | 5 | 0.20 | 1 | 1 | 0 |
| ....................................................................................................CTTCTCCGTCACTCAAACCTACT....................................... | 23 | 0 | 5 | 0.20 | 1 | 1 | 0 |
| ......................................................................................................TCTCCGTCACTCAAACCTACTTGAGTAT................................ | 28 | 0 | 5 | 0.20 | 1 | 1 | 0 |
| .........................................................................................................CCGTCACTCAAACCTACTTGAGTATTTC............................. | 28 | 1 | 5 | 0.20 | 1 | 0 | 1 |
| .........................................................................................................CCGTCACTCAAACCTACTTGAGTATTT.............................. | 27 | 0 | 5 | 0.20 | 1 | 0 | 1 |
| .......................................................................................................CTCCGTCACTCAAACCTACTTGAGTAT................................ | 27 | 0 | 5 | 0.20 | 1 | 1 | 0 |
| .......................................................................................................CTCCGTCACTCAAACCTGCTTGAGTATTT.............................. | 29 | 1 | 5 | 0.20 | 1 | 1 | 0 |
| .......................................................................................................CTCCGTCACTCAAACCTACTTGAGTATTT.............................. | 29 | 0 | 5 | 0.20 | 1 | 1 | 0 |
| .........................................................................................................CCGTCACTCAAACCTACTTGAGTATT............................... | 26 | 0 | 5 | 0.20 | 1 | 0 | 1 |
| ...................................................................................................GCTTCTCCGTCACTCAAACCTACT....................................... | 24 | 0 | 5 | 0.20 | 1 | 0 | 1 |
| .....................................................................................................TTCTCCGTCACTCAAACCTACTT...................................... | 23 | 0 | 5 | 0.20 | 1 | 0 | 1 |
| .................................................................................................CTGCTTCTCCGTCACTCAAACCTAC........................................ | 25 | 0 | 5 | 0.20 | 1 | 1 | 0 |
| .........................................................................................................CCGTCACTCAAACCTACTTGAGTAT................................ | 25 | 0 | 5 | 0.20 | 1 | 1 | 0 |
| .............................................................................................AGGTCTTCTTCTCCGTAACTCAAACCT.......................................... | 27 | 2 | 5 | 0.20 | 1 | 0 | 1 |
| .....................................................................................................TTCTCCGTCACTCAAACCTACT....................................... | 22 | 0 | 5 | 0.20 | 1 | 0 | 1 |
| ..........................................................................................................CGTCACTCAAACCTACTTGAGTATT............................... | 25 | 0 | 5 | 0.20 | 1 | 1 | 0 |
| .........................................................................................................CCGTCACTCAAACCTACTTGAGTATTC.............................. | 27 | 1 | 5 | 0.20 | 1 | 1 | 0 |
| .........................................................................CATTCGTTAGCTCTAATAATAGGT................................................................. | 24 | 1 | 5 | 0.20 | 1 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6500:14037522-14037683 + | dmo_324 | TTTT---AGATACGGACTAAGTCGTAACAACAAATGCTGCTACGAGTACACATTGCCTCCTAAATGGATGCAAGCAGTAAGCAATCGAGCTTATTATC---------CAGACGAAGAGGCA---G--------------------TGAGTTTGGATGAACT----CATAAATGAGCTGCAGGA-----GTAATCAAAT--TTA----------ACATT |
| droWil2 | scf2_1100000004542:78476-78630 - | TTTC---AGATACGGACTTAATCGTAGCACCAAATGCTGCTATGATTACATTTTACCCCAAAAATTGTTGCGTGCAGTGAGCAACCGATCTTATTATC---------CTGACGAGGAAGAA---G--------------------TAAATTTAAATGGGAT----ATTAAGTGAAATATAAAA-----ATA---------AAA----------AAATT | |
| dp5 | 3:15793761-15793816 - | TTTT-ACAGATATGGATTGCGCCGTCATGACAAATGTTGCCCCAACTACCAATTACC----------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_89:49192-49254 + | ATA----------GGGCTGCGGAGACAAGGAAAATGTTGCAACAACTAAGAATTGCCAGTTAAATATTTGAAT------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_13417:2714488-2714653 + | -------AGATTCGGATTGCGTCGCCCTAAATTATGTTGTCATAATTATAACTTGCCTAAAAAGTATCTCCAATTAATTGGCTCCATGGCTACTTATG---------AGGACA------GCAATAATGATGTAGGGGAA------AAATAATCAAAGATGC----CTTAAAACTATTATA--------TTAATTAAAA--AAA----------AAAAT | |
| droBip1 | scf7180000394194:1588-1761 - | TTGT---AGATACGGATTACGCCGAGCAAACAAATGCTGTTACGATTATCAAGTGCCAGCAGAATGCCTTAACAAGATTGGAACGAATGAGTCGTATA---AGAATTCAGCTGCAGAAGAA---A--------------------AAACTATTCACGAACTTGATTACGAAAATGCTAAATAA-----CTAATAAAAATTTTG----------ATTTT | |
| droKik1 | scf7180000298547:8183-8349 + | TTTT---AGGTATGGACTACGCCGAAGCAATAAATGCTGTTTTGACTACCAGCTACCAAGTAGCTGCGTTGAGAGACTCATCCAACGGGATTATTATC---------CGTCCGAAGATGAT---G--------------------AGCATTGCGATGATAT----ATTAAATATAATTTAAATATAATTTCTTTATAT--ATA----------ATGTC | |
| droFic1 | scf7180000453976:4379-4539 + | TTTT---AGAAAGGGGCTGCGTCGACACAATAAGTGCTGTTTTAACTACAATCTACCACCCATCTGCCTAACTACAATTGAACAAAAGAATTTTTATC---------CAAGCGATGATGTC---G--------------------GTAATACTGACGATAT----CTCAAATATACTATAATT-----CT-------------TAATTTTTGTACATT | |
| droEle1 | scf7180000491107:281689-281834 + | TTTA---AGATATGGTCTGCGTTGAAGCAATAAATGGTGTTTTGATTACCAGCTGCCAAGCAGCTGCTCAGCCACACTAGTCCAACAGGATTATTGTC---------CGTCCGAAGAAGGA---G--------------------AACATTTGGACGACGA----ATAAAATATAATTTA---------------------------------ACTTT | |
| droRho1 | scf7180000776640:744-903 - | TTTT---ATATACAGAA------GGTTTGTAAAATGCTGCGTCGACTATGAAGTACCTGTAAAATATCTGCGCATGATAGGCACTAATATGTCATACG---------CAAATAAGGACGACCAAG--------------------AAGATATTGATAAAAT----TATGGAAATATTTTAAGAAA---AA-------------TTATCTTAATAAATT | |
| droBia1 | scf7180000302291:1506997-1507157 - | ATTT---TAATATGGGCTTCGTCGACACAATAAGTGCTGTTTTAACTACAGTATACCACCCAGCTGCCTAAATAAAATGGAACAAAAGCATTTTTACC---------AAACCGATGATGCC---G--------------------ATAATGTTGACGATAT----CTTAAATGTCCTATAAGT-----TT-------------TAACTTTTGTACATT | |
| droTak1 | scf7180000414953:1177-1326 + | -------AGATTTGGCCTACGACGACACGGATCATGCTGCCACAATTACAATATACCTAATAAGTACCTTAAAATGATTGGTACCAAAGCTGCATACTCCGAAGAGGCAACCGAGGATGAC---G--------------------AAATTATTGATTCTGC----CATTAAACTAATCT----------------------------------AATTT | |
| droEug1 | scf7180000409859:44033-44173 + | TTTT---AGGTACGGGCTGCGTCGACACAATAAGTGCTGTTTTAACTATAATATACACCCCAGCTGCCTATCTACAATTGAAGACAAGAAATTTTATC---------CAACTGATGATGAT---G--------------------ATAATATTGACGACAT----CTTAAATATAATATA-------------------------------------- | |
| droYak3 | 2L:17644214-17644381 - | TTTTTACAAATATGGTTTGCGCAGAGAAAATATATATTGTCACAGTTATATAGGTCCTCCAAATTACCTAAAACTTATTGGAACTAACCAAAACTATA---------TAAATGAAAAGGCA---A--------------ACTATGCAGATATGGATGAAAG----CAC---TATATTTTGGGA-----GTCATTTTAA--TAT----------AGACT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/15/2015 at 03:11 PM