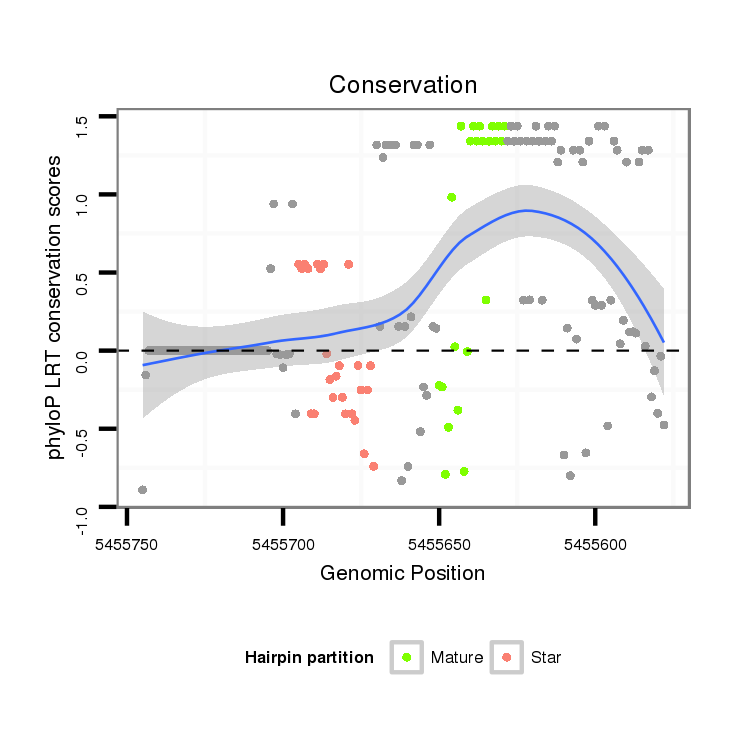
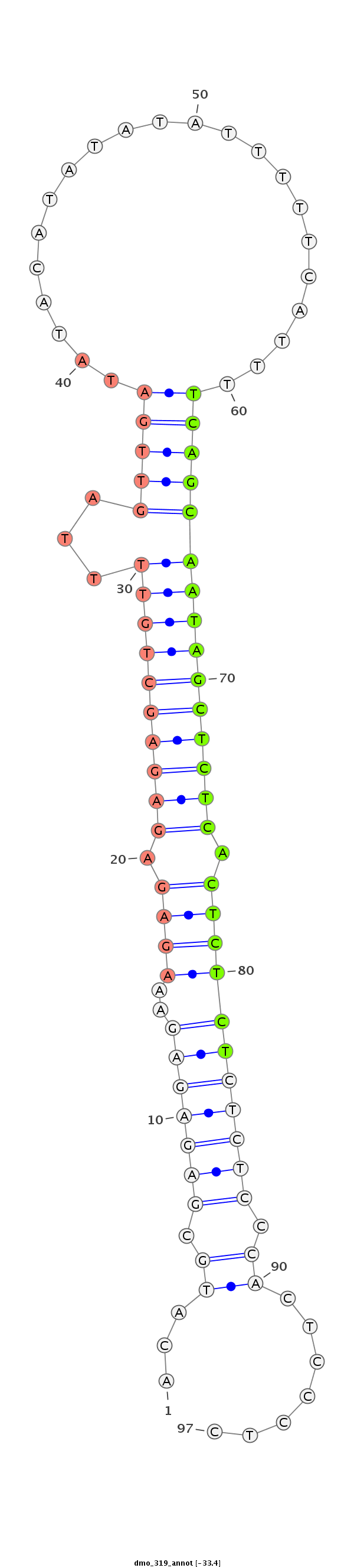
| ...............................................................................................TCAGCAATAGCTCTCACTCTCT................................................... |
22 |
0 |
1 |
117.00 |
117 |
89 |
20 |
5 |
2 |
0 |
1 |
| ...............................................................................................TCAGCAATAGCTCTCACTCTCC................................................... |
22 |
1 |
1 |
29.00 |
29 |
28 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................................................TCAGCAATAGCTCTCACTCTC.................................................... |
21 |
0 |
1 |
9.00 |
9 |
5 |
2 |
1 |
0 |
0 |
1 |
| ..................................................AGAGAGAGAGCTGTTTTAGTTGATA............................................................................................. |
25 |
0 |
1 |
8.00 |
8 |
4 |
0 |
0 |
0 |
4 |
0 |
| ...............................................................................................TCAGCAATAGCTCTCACTCTCTC.................................................. |
23 |
0 |
1 |
6.00 |
6 |
2 |
4 |
0 |
0 |
0 |
0 |
| ...............................................................................................TGAGCAATAGCTCTCACTCTCT................................................... |
22 |
1 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................CAGCAATAGCTCTCACTCTCTC.................................................. |
22 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................AGCAATAGCTCTCACTCTCTCC................................................. |
22 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................AGCAATAGCTCTCACTCTCTCT................................................. |
22 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................CCAGCAATAGCTCTCACTCTCT................................................... |
22 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
| ..................................................AGAGAGAGAGCTGTTTTAGTTG................................................................................................ |
22 |
0 |
1 |
2.00 |
2 |
1 |
0 |
0 |
1 |
0 |
0 |
| ...............................................................................................TCAGCAATAGCTCTCTCTCTCT................................................... |
22 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................TCAGCAATAGCTCTCACTCT..................................................... |
20 |
0 |
1 |
2.00 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
| ..................................................AAAGAGAGAGCTGTTTTAGTTGATA............................................................................................. |
25 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................TCAGCACTAGCTCTCACTCTCT................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| .................................................AAGAGAGAGAGCTGTTTTAGTTGATA............................................................................................. |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ......AGATCGATGCAGCTGATGGATC............................................................................................................................................ |
22 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .................................ATACATGCGGGAGAGAAAG.................................................................................................................... |
19 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| ...............................GGATACATGCGAGAGAGAA...................................................................................................................... |
19 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
| .................................ATACATGCGAGAGAGAAAG.................................................................................................................... |
19 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
| ..................................................ATAGAGAGAGCTGTTTTAGTTGATA............................................................................................. |
25 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................TCAGCAATAGCTCTCACTCTCTCT................................................. |
24 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...............................................................................................TGAGCAATAGCTCTCACTCTC.................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................GATCTCAGCAATAGCTCTCACTCTCT................................................... |
26 |
3 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................GCAGCAATAGCTCTCACTCTCT................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................TCAGCAATAGCTCTCACGCTCT................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................CCAGCAATAGCTCTCACTCTC.................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................ATCAATAGCTCTCACTCTCTC.................................................. |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................TGAGCAATAGCTCTCACTCTCC................................................... |
22 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................TCAGCGATAGCTCTCACTCTCT................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................TGAGCAATAGCTATCACTCTCT................................................... |
22 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................TTAGCAATAGCTCTCACTCTCT................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| .................................................AAGAGAGAGAGCTGTTTTAGTTGA............................................................................................... |
24 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...............................................................................................TCAGCAATAGCTTTCACTCTC.................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................CTCAGCAATAGCTCTCACTCTCT................................................... |
23 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................TCAGCAATAGCTCTAACTCTCC................................................... |
22 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................CAGCAATAGCTCTCACTCTC.................................................... |
20 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................AGCAATAGCTCTCACTCTCTC.................................................. |
21 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................TCAATAGCTCTCACTCTCT................................................... |
19 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...........GTTGCAGGTGATGGATCA........................................................................................................................................... |
18 |
2 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |