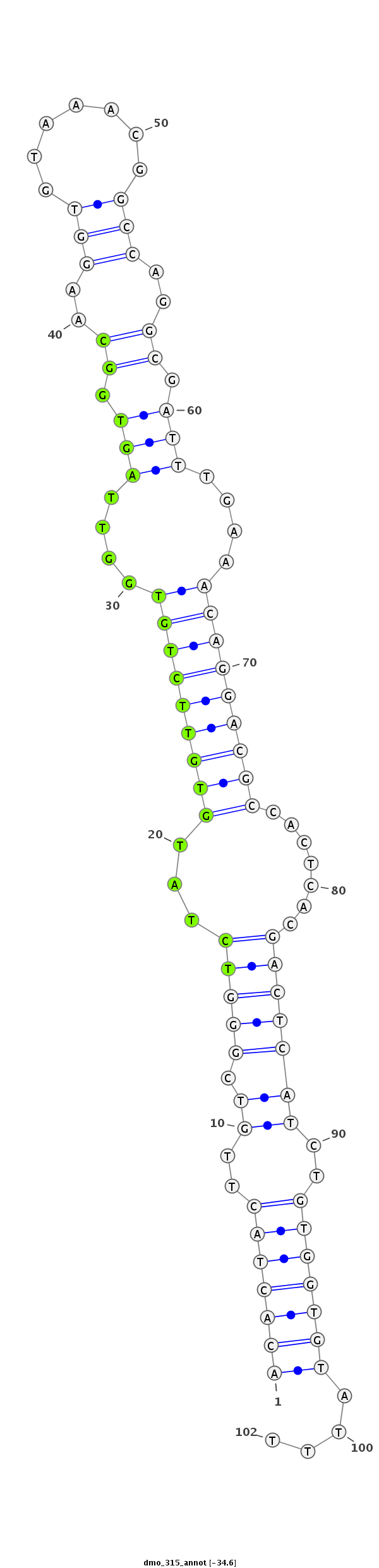
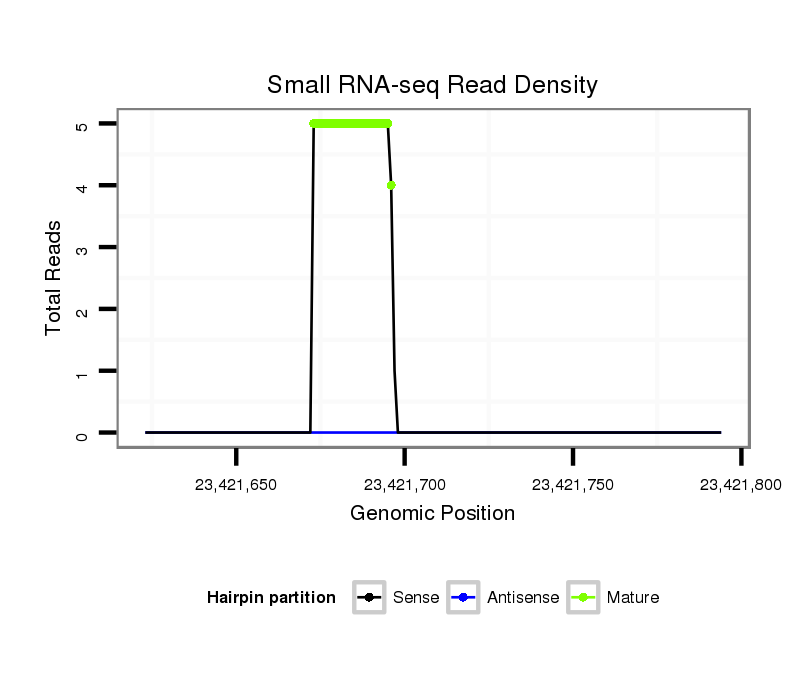
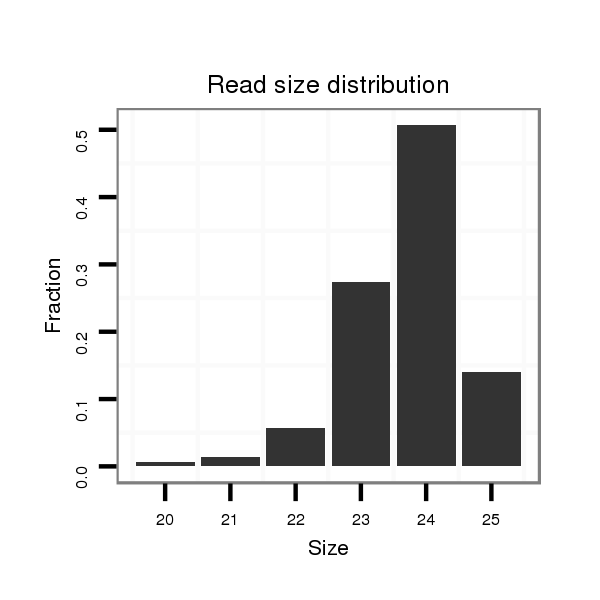
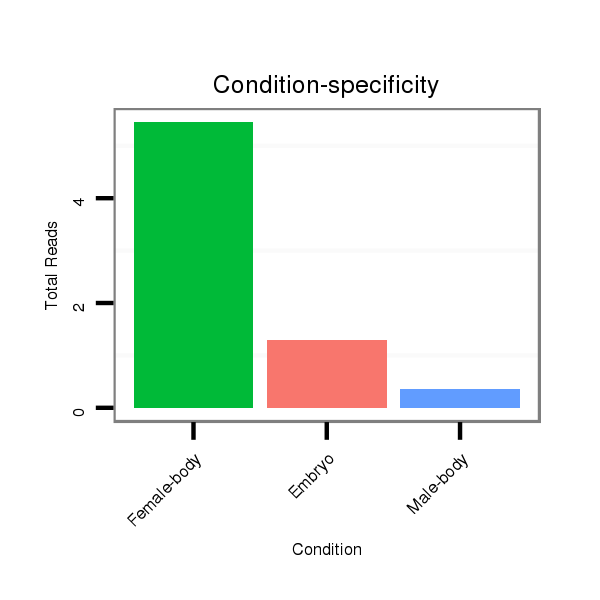
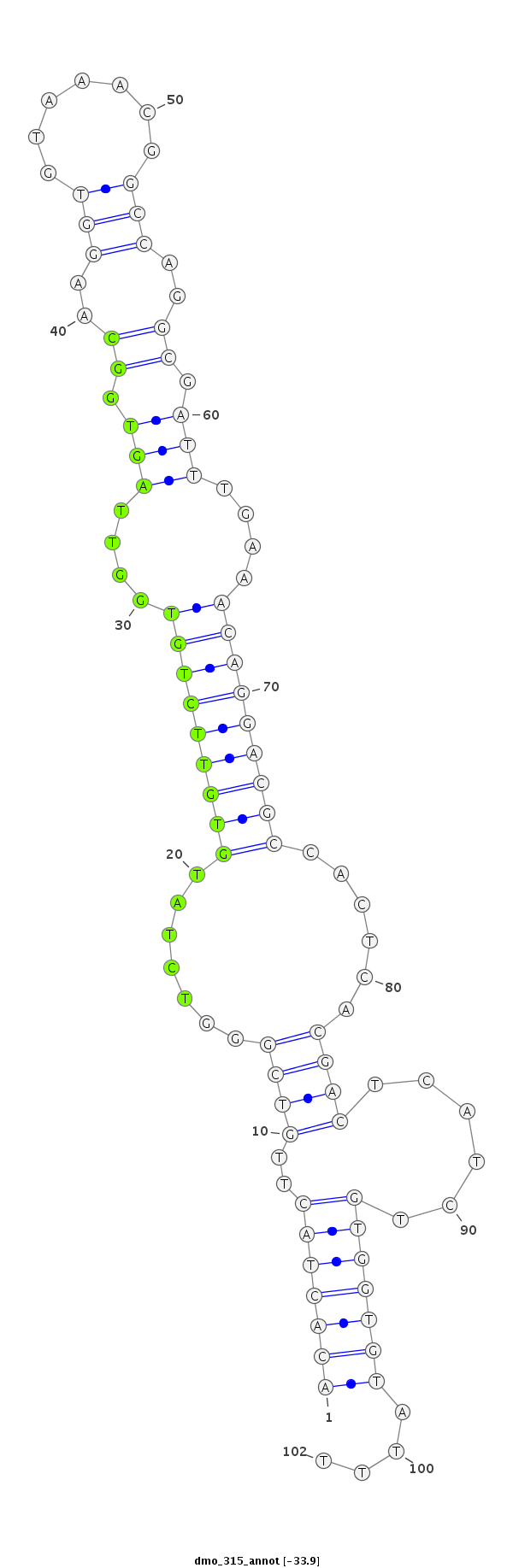
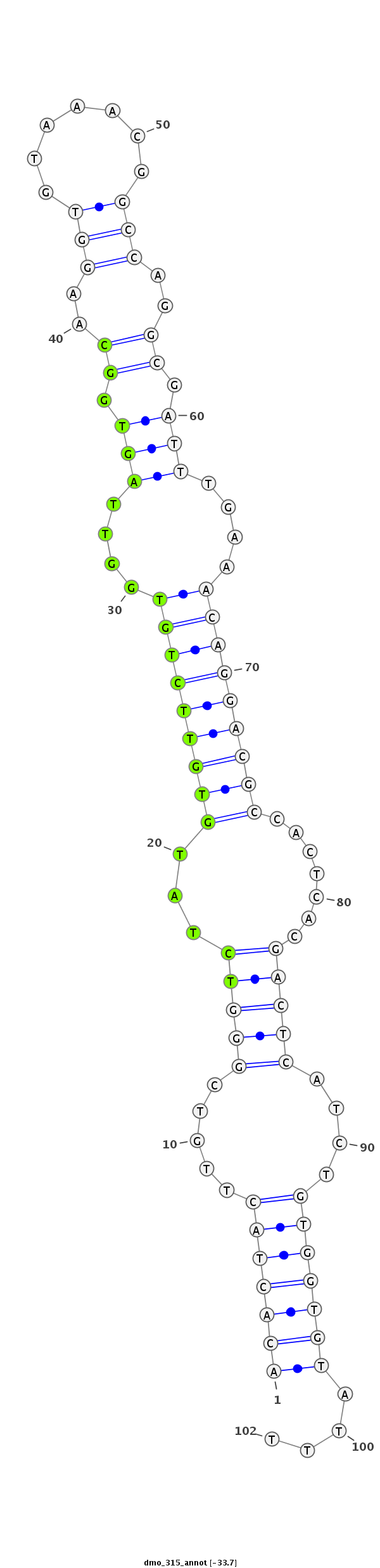
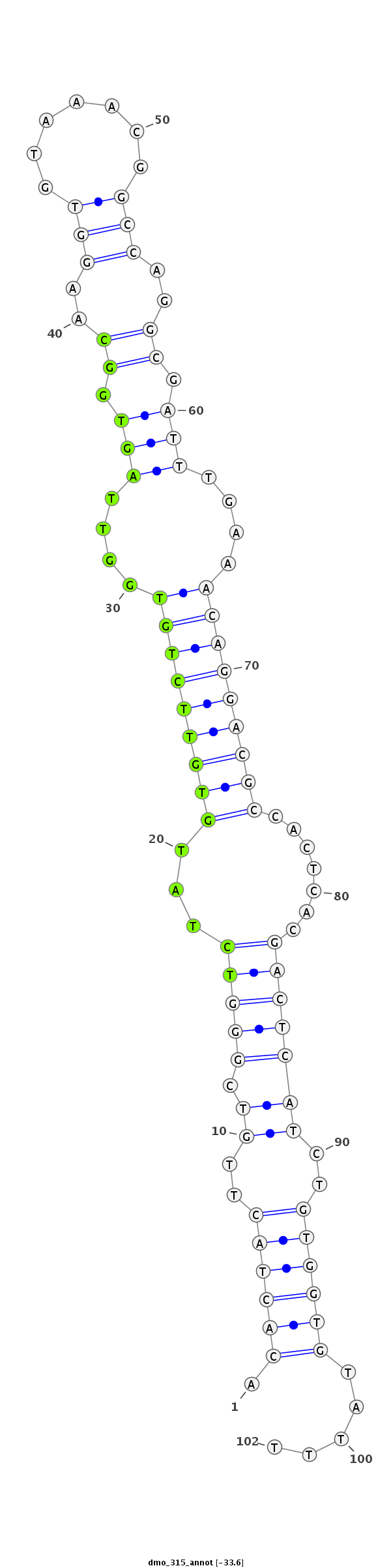
| ..................................................TCTATGTGTTCTGTGGTTAGTGGC.................................................................................................. |
24 |
0 |
5 |
3.60 |
18 |
13 |
4 |
0 |
1 |
| ..................................................TCTATGTGTTCTGTGGTTAGTGG................................................................................................... |
23 |
0 |
20 |
1.55 |
31 |
19 |
5 |
7 |
0 |
| ..................................................TCTATGTGTTCTGTGGTTAGTGGCA................................................................................................. |
25 |
0 |
2 |
1.00 |
2 |
2 |
0 |
0 |
0 |
| ...................................................CTATGTGTTCTGTGGTTAGTGGC.................................................................................................. |
23 |
0 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
| ...................................................CTATGTGTTCTGTGGTTAGTGG................................................................................................... |
22 |
0 |
20 |
0.20 |
4 |
3 |
1 |
0 |
0 |
| ..................................................TCTATGTGTTCTGTGGTTAGTG.................................................................................................... |
22 |
0 |
20 |
0.20 |
4 |
4 |
0 |
0 |
0 |
| .................................................GTCTATGTGTTCTGTGGTTAGTGGC.................................................................................................. |
25 |
0 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
| ......................................................TGGGTTCTGGGGGTAGTGGC.................................................................................................. |
20 |
3 |
10 |
0.10 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TCTATGTGTTCTGTGGTTAGT..................................................................................................... |
21 |
0 |
20 |
0.10 |
2 |
2 |
0 |
0 |
0 |
| ...............AAAAGTGTGCAAAACGCTTTACACTAC.................................................................................................................................. |
27 |
0 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| .................................................GTCTATGTGTTCTGTGGTTAGTGG................................................................................................... |
24 |
0 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TCTATGTGTTCTGTGGTTAG...................................................................................................... |
20 |
0 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ...................GTGTGCAAAACGCTTTACACT.................................................................................................................................... |
21 |
0 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ..................................................TCTATGTGTTCTGTGGTTAGTGGA.................................................................................................. |
24 |
1 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| ........................................................................GCTAGGGGTAAAAGGCCA.................................................................................. |
18 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ..................AGTGTGCAAAACGCTTTACACTACT................................................................................................................................. |
25 |
0 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |