ID:dmo_302 |
Coordinate:scaffold_6540:11863782-11863845 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -44.1 | -43.9 | -43.5 |
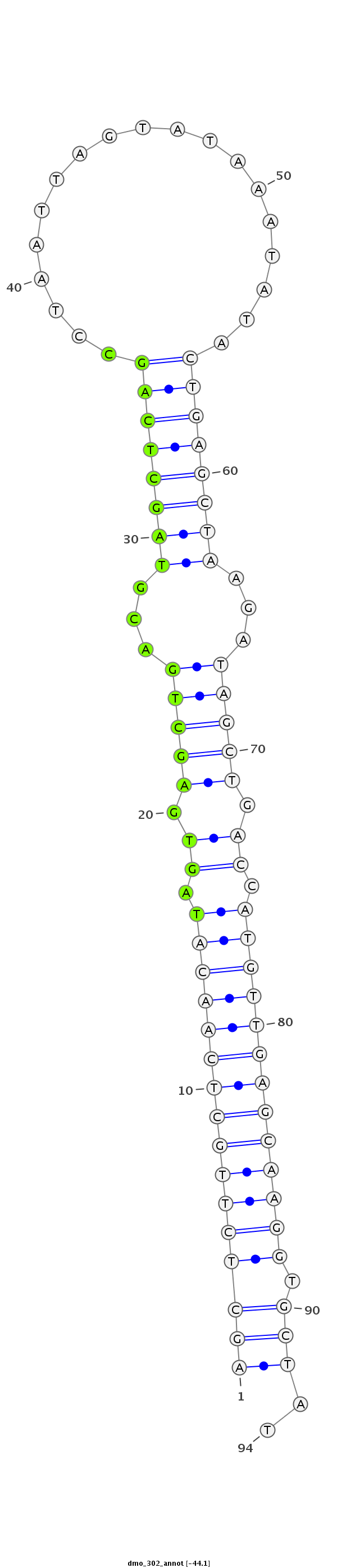 |
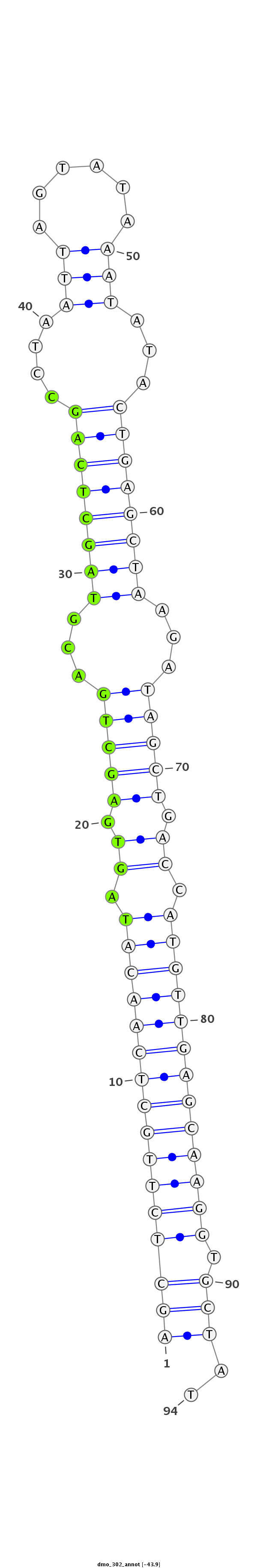 |
 |
CDS [Dmoj\GI23140-cds]; exon [dmoj_GLEANR_8297:1]; intron [Dmoj\GI23140-in]
No Repeatable elements found
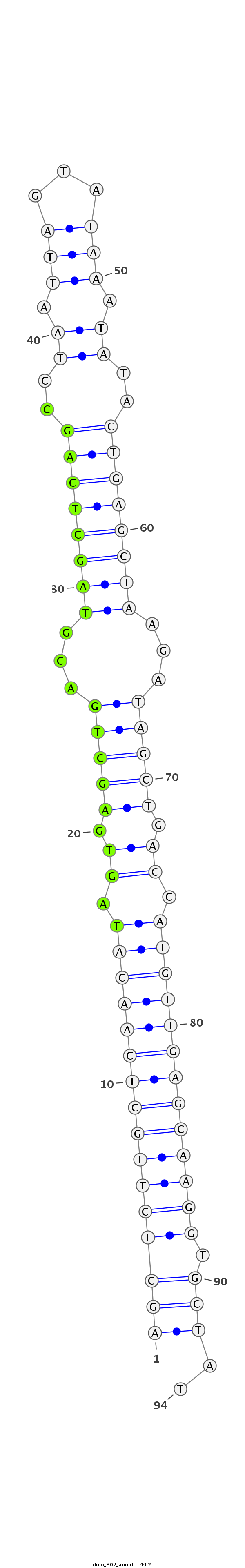
| --------------------------------------------------------------------------------------------------------------###################----------------------------------- TGAATTGTAAGAAGTTGTTCAACTCTGGCCCAGTCAGCTCTTGCTCAACATAGTGAGCTGACGTAGCTCAGCCTAATTAGTATAAATATACTGAGCTAAGATAGCTGACCATGTTGAGCAAGGTGCTATGTATGTCATATTATATATGCTATGCTATGCTATAT ***********************************((((((((((((((((.((.(((((...((((((((..((.(((...))).))..))))))))...))))).)).))))))))))))).)))..*********************************** |
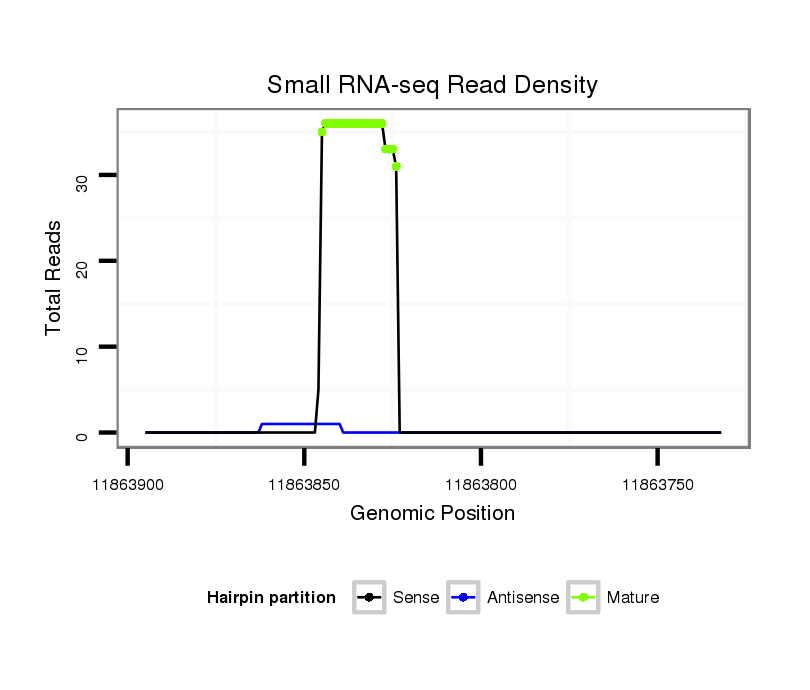
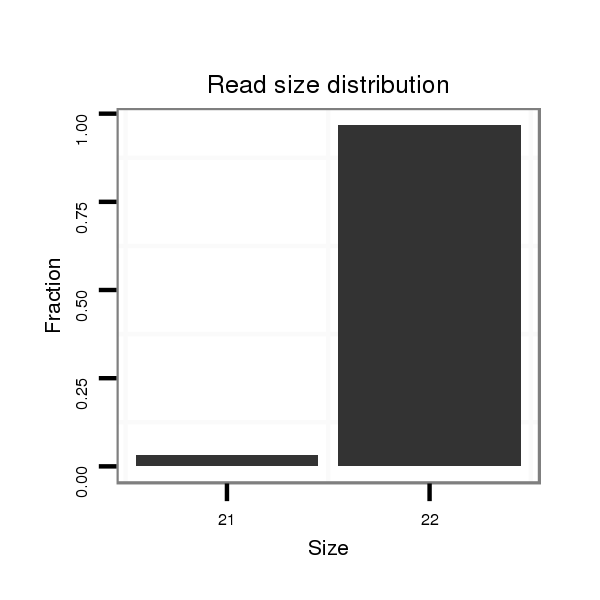
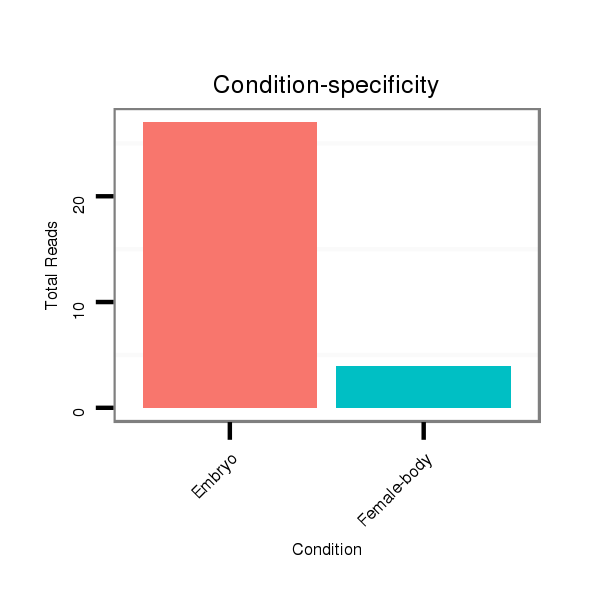
Read size | # Mismatch | Hit Count | Total Norm | Total | M060 embryo |
M046 female body |
V041 embryo |
V056 head |
|---|---|---|---|---|---|---|---|---|---|
| ..................................................TAGTGAGCTGACGTAGCTCAGC............................................................................................ | 22 | 0 | 1 | 30.00 | 30 | 25 | 4 | 1 | 0 |
| .................................................ATAGTGAGCTGACGTAGCT................................................................................................ | 19 | 0 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 |
| .................................................ATAGTGAGCTGACGTAGCTCAG............................................................................................. | 22 | 0 | 1 | 2.00 | 2 | 0 | 1 | 0 | 1 |
| ..................................................TAGTGAGCTGACGTAGCTCATC............................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................TAGTGAGCTGACGCAGCTCAGC............................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................TGGTGAGCTGACGTAGCTCAGC............................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...................................................AGTGAGCTGACGTAGCTCAGC............................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
|
ACTTAACATTCTTCAACAAGTTGAGACCGGGTCAGTCGAGAACGAGTTGTATCACTCGACTGCATCGAGTCGGATTAATCATATTTATATGACTCGATTCTATCGACTGGTACAACTCGTTCCACGATACATACAGTATAATATATACGATACGATACGATATA
***********************************((((((((((((((((.((.(((((...((((((((..((.(((...))).))..))))))))...))))).)).))))))))))))).)))..*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V056 head |
|---|---|---|---|---|---|---|
| .................................AGTCGAGAACGAGTTGTATCACT............................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6540:11863732-11863895 - | dmo_302 | TGAATTGTAAGAAGTTGTTCAACTCTGGCCCAGTCAGCTCTTGCTCAACATAGTGAGCTGACGTAGCTCAGCCTAATTAGTATAAATATACTGAGCTAAGATAGCTGACCATGTTGAGCAAGGTGCTATGTATGTCATATTATATATGCTATGCTATGCTATAT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
Generated: 05/15/2015 at 03:06 PM