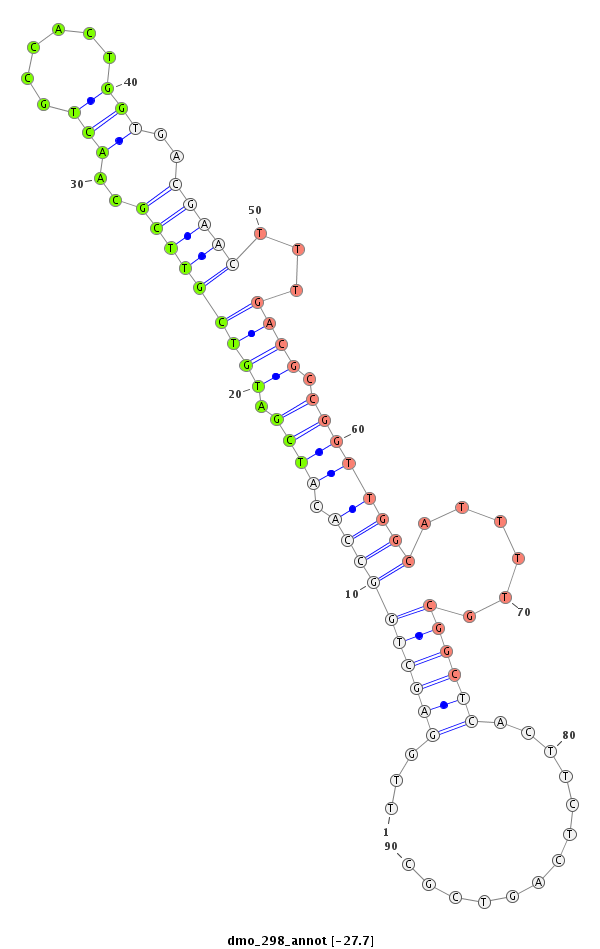
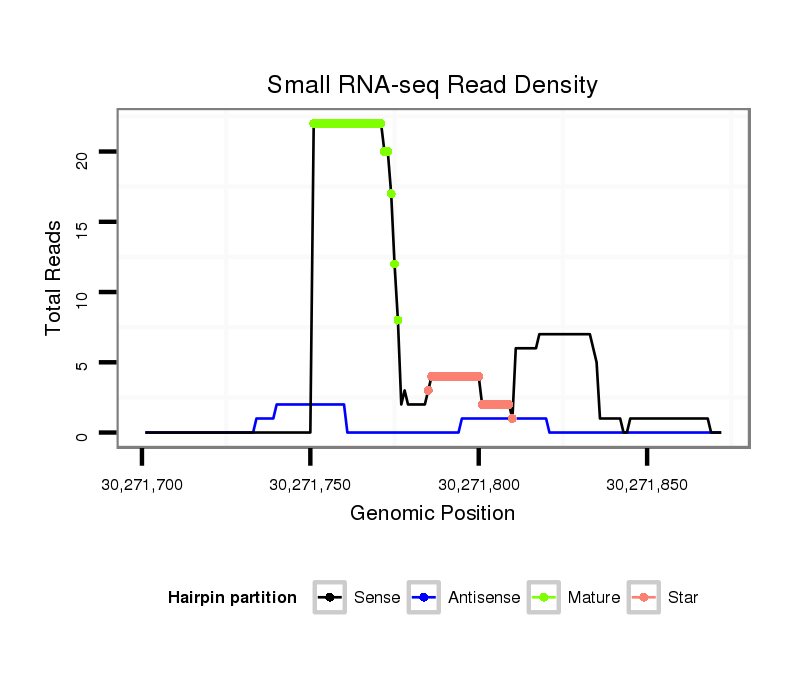
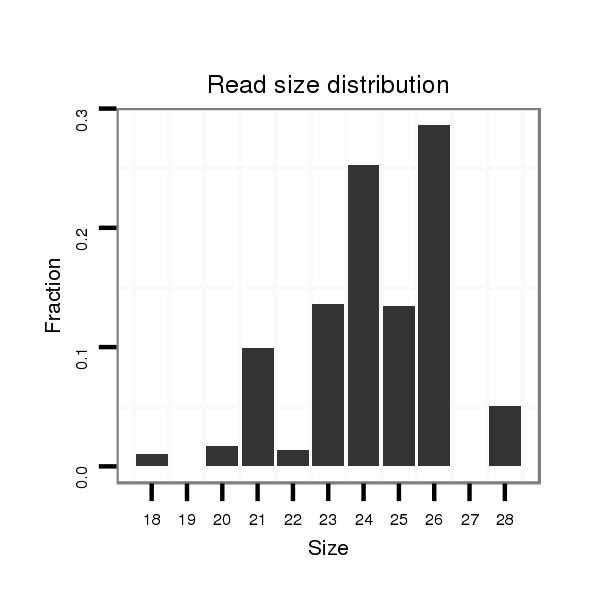
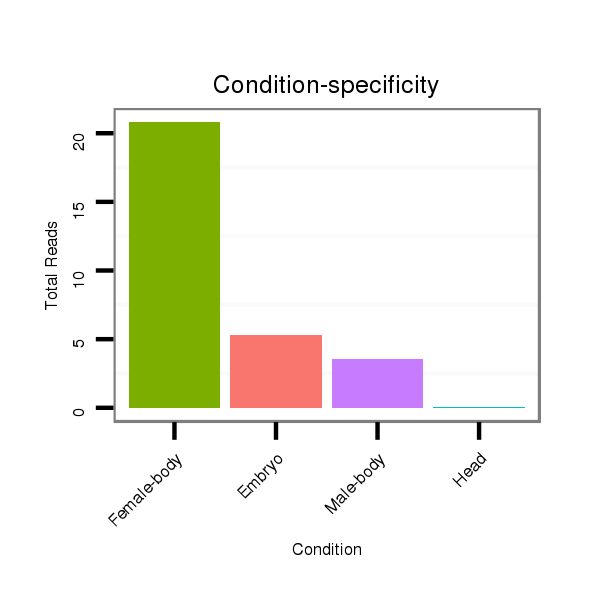
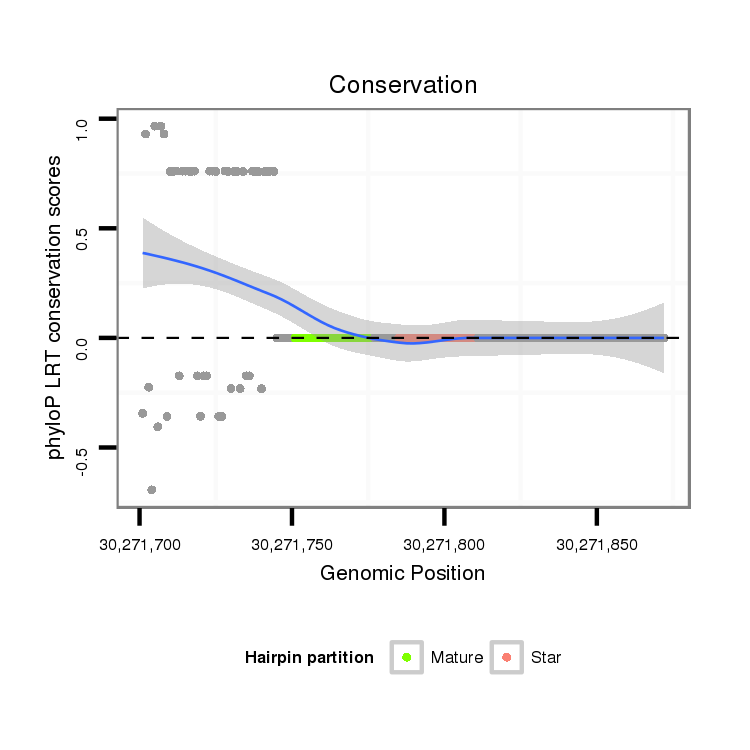
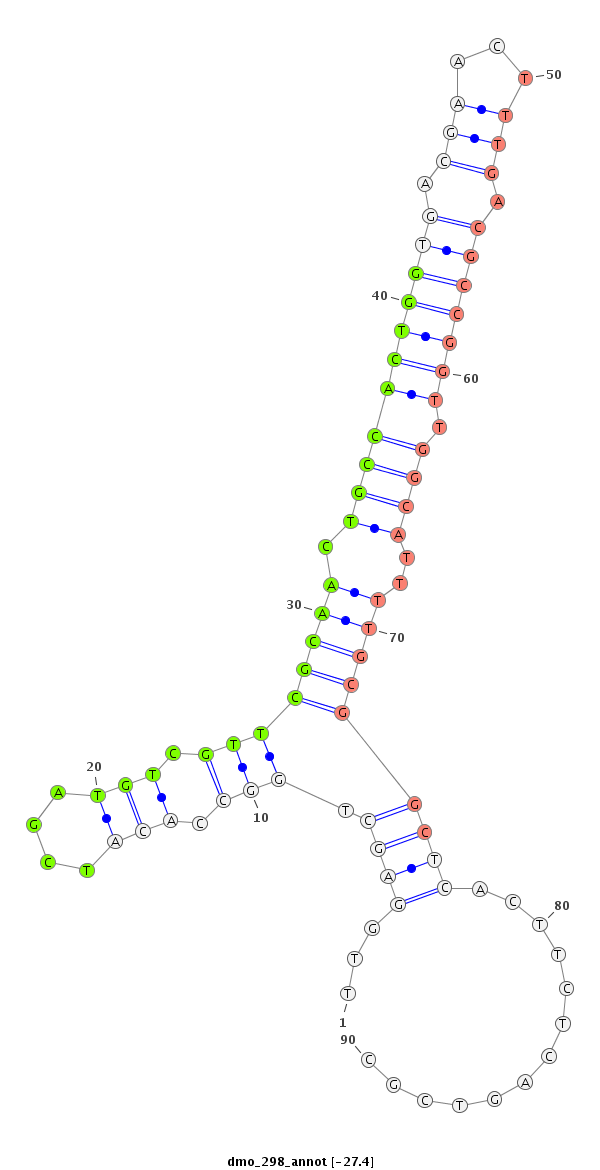
ID:dmo_298 |
Coordinate:scaffold_6500:30271751-30271822 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -27.4 | -27.4 | -27.3 |
 |
 |
 |
intergenic
No Repeatable elements found
| mature | star |
|
CCGTTTAGCGCATTTTGTCCCCTTCGGACTGCTGTTTGGAGCTGGCCACATCGATGTCGTTCGCAACTGCCACTGGTGACGAACTTTGACGCCGGTTGGCATTTTGCGGCTCACTTCTCAGTCGCGCACAGCTCCCAACAGCAATCCGAAATTTGTCTGCAGTCAGCTTAAA
***********************************...((((((((((.((((.(((((((((..(((......)))..)))))...)))).))))))))......)))))).............*********************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V110 male body |
V049 head |
V041 embryo |
V056 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................TCGATGTCGTTCGCAACTGCCACTGG................................................................................................ | 26 | 0 | 2 | 7.50 | 15 | 9 | 1 | 5 | 0 | 0 | 0 |
| ..................................................TCGATGTCGTTCGCAACTGCCACT.................................................................................................. | 24 | 0 | 2 | 5.50 | 11 | 5 | 5 | 1 | 0 | 0 | 0 |
| ................................................................................GAACTTTGGGCCCGGTTGGC........................................................................ | 20 | 3 | 7 | 4.43 | 31 | 21 | 1 | 8 | 1 | 0 | 0 |
| ..................................................TCGATGTCGTTCGCAACTGCCACTG................................................................................................. | 25 | 0 | 2 | 4.00 | 8 | 7 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................TCACTTCTCAGTCGCGCACAGCTCC..................................... | 25 | 0 | 1 | 4.00 | 4 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..................................................TCGATGTCGTTCGCAACTGCCAC................................................................................................... | 23 | 0 | 20 | 3.05 | 61 | 61 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TCGATGTCGTTCGCAACTGCC..................................................................................................... | 21 | 0 | 20 | 2.95 | 59 | 13 | 46 | 0 | 0 | 0 | 0 |
| ..................................................TCGATGTCGTTCGCAACTGCCACTGGTG.............................................................................................. | 28 | 0 | 2 | 1.50 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TCACTTCTCAGTCGCGCACAGCTC...................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................TTTGACGCCGGTTGGCATTTTGCGGC.............................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................TTGACGCCGGTTGGCATTTTGCGG............................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................CAACTGCCGTTGGTGACGAA......................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............TTTGTCCCCTTCAAACCGCTGTT........................................................................................................................................ | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................TCACTTCTCAGTCGCGCACATCTCCC.................................... | 26 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TCCGAAATTTGTCTGCAGTCAGCT.... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TCATTTCTCAGTCGCGCACAGCTCC..................................... | 25 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TCACTTCTCAGTCGCGCACAGCT....................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................GACGAACTTTGACGCCGGTTGGC........................................................................ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TCACTTCTCAGTCGCGCACTGCTC...................................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................TGACGAACTTTGACGCCGGTTGGC........................................................................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................TCAGTCGCGCACAGCTCCCAACAGC.............................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TCGATGTCGTTCGCAACTGCCACTTAT............................................................................................... | 27 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TCGATGTCGTTCGCAACTGCCATTG................................................................................................. | 25 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TCGATGTCGTTCCCAACTGCCACTGG................................................................................................ | 26 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................CAACTGCCGTTGGTGACGA.......................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............TTGGTCCCCTTCGAACAGCTGTT........................................................................................................................................ | 23 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TCGATGTCGTTCGCAACTGC...................................................................................................... | 20 | 0 | 20 | 0.45 | 9 | 8 | 0 | 0 | 1 | 0 | 0 |
| ...................................................CGATGTCGTTCGCAACTGCCAC................................................................................................... | 22 | 0 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TCGATGTCGCTCGCAACTGCCACTGGT............................................................................................... | 27 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TCGATGTCGTTCGCAACTGTCACTT................................................................................................. | 25 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TCGATGTCGTTCGCAACT........................................................................................................ | 18 | 0 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................GAACTTTGGGCCCGGTTGG......................................................................... | 19 | 3 | 17 | 0.29 | 5 | 3 | 0 | 2 | 0 | 0 | 0 |
| ..................................................TCGATGTCGTTCGCAACTGCCACCGGTGA............................................................................................. | 29 | 1 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ..............TTGTCCCCTTCACACAGCTGT......................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................TGGCCACATCGATGTCGTTCGCAAC......................................................................................................... | 25 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TCGTTGTCGTTCGCAACTGCCAC................................................................................................... | 23 | 1 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| .............TTGGTCCCCTTCGCCCTGCTGT......................................................................................................................................... | 22 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................TCGATGTCGTTCGCAACTGCCAT................................................................................................... | 23 | 1 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................GCCACATCGATGTCGTTCGCAACT........................................................................................................ | 24 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..........................................TGGCCACATCGATGTCGTTCGCAA.......................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................GATGTCGTTCGCAACTGCCA.................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TCGTTGTCGTTCGCAACTGCCA.................................................................................................... | 22 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................GCCACATCGATGTCGTTCGCAACTGCC..................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................TGGCCACATCGATGTCGTTCGCAACTG....................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TCGATGTCGTTCGCAACTGCCA.................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................TGGCCACATCGATGTCGTTCGCAACT........................................................................................................ | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................TGGATGTCGTTCGCAACTGCC..................................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................CCACATCGATGTCGTTCGCAAC......................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................CATCGATGTCGTTCGCAACTGCCAC................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TCGATGTCGTTCGCAACTCCACC................................................................................................... | 23 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TCCATGTCGTTCGCAACTGCC..................................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................TGGCCACATCGATGTCGTTCGCAACTGCC..................................................................................................... | 29 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
|
GGCAAATCGCGTAAAACAGGGGAAGCCTGACGACAAACCTCGACCGGTGTAGCTACAGCAAGCGTTGACGGTGACCACTGCTTGAAACTGCGGCCAACCGTAAAACGCCGAGTGAAGAGTCAGCGCGTGTCGAGGGTTGTCGTTAGGCTTTAAACAGACGTCAGTCGAATTT
***********************************************...((((((((((.((((.(((((((((..(((......)))..)))))...)))).))))))))......)))))).............*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V041 embryo |
|---|---|---|---|---|---|---|---|---|
| .......................................TCGACCGGTGTAGCTACAGCA................................................................................................................ | 21 | 0 | 20 | 1.50 | 30 | 30 | 0 | 0 |
| ..............................................................................................CAACCGTAAAACGCCGAGTGAAGAGT.................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................CAAACCTCGACCGGTGTAGCTACAGCA................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..CAAATCGCGTAAAACAGGAGAAGCCT................................................................................................................................................ | 26 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ................................................................................................................................................CGGCTTTAAACAGACGTCAGTCGAATTT | 28 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................CAAACCTCGACCGGTTTAGCTACAGCA................................................................................................................ | 27 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| .................................................................................................................................TCGAGGGTTGTCGTTAGGCTTT..................... | 22 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| .....................................CCTCGACCGGTGTAGCTACAGGA................................................................................................................ | 23 | 1 | 20 | 0.25 | 5 | 0 | 5 | 0 |
| ..................................CGACCTCGACCGGTGTAGCTACAGCA................................................................................................................ | 26 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| ..................................................................................................................................CGACCGGTGTCGTTAGGCTTT..................... | 21 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 |
| .......................................TCGACCGGTGTAGCTACACCA................................................................................................................ | 21 | 1 | 20 | 0.10 | 2 | 0 | 2 | 0 |
| .....................................CCTCGACCGGTGTAGCTACGGCA................................................................................................................ | 23 | 1 | 20 | 0.10 | 2 | 0 | 2 | 0 |
| .....................................CCTCGACCGTTGTAGCTACAGCA................................................................................................................ | 23 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| .......................................TCGACCGTTGTAGCTACAGCA................................................................................................................ | 21 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| .....................................CCTCGTCCGGTGTAGCTACAGCA................................................................................................................ | 23 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ..............................................GTGTAGCTACAGCAAGCGTTGACG...................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ........................................CGACCGGTGTAGCTACGGCACT.............................................................................................................. | 22 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ......................................CTCGACCGGTGTAGCTACAGCA................................................................................................................ | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| .....................................CCTCGACCGGTGTAGCTACAGCG................................................................................................................ | 23 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ......................................CTCGACCGGTGTAGCTACAGTA................................................................................................................ | 22 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| .......................................TCGACCGGTGTAGCTGCAGCA................................................................................................................ | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .........................................GACCGGTGTAGCTACAGCA................................................................................................................ | 19 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| .....................................ACTCGACATGTGTAGCTACAGCA................................................................................................................ | 23 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ........................................CGACCGGTGTAGCTACAGCAAGCGTGG......................................................................................................... | 27 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ........................................CGACCGGTGTAGATACAGCA................................................................................................................ | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6500:30271701-30271872 + | dmo_298 | CCGTTTAGCGCATTTTGTCC-CCTTCGGACTGCTGTTTGGAGCTGGCCACATCGATGTCGTTCGCAACTGCCACTGGTGACGAACTTTGACGCCGGTTGGCATTTTGCGGCTCACTTCTCAGTCGCGCACAGCTCCCAACAGCAATCCGAAATTTGTCTGCAGTCAGCTTAAA |
| droTak1 | scf7180000414536:205388-205432 + | TCGATTAGAGCACTTTGTTAGTTTTCTTACAGCAGCCTGGTGCTG-------------------------------------------------------------------------------------------------------------------------------- | |
| droYak3 | 3R:16440777-16440784 + | CCAGTCAG--------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 05/15/2015 at 03:07 PM