
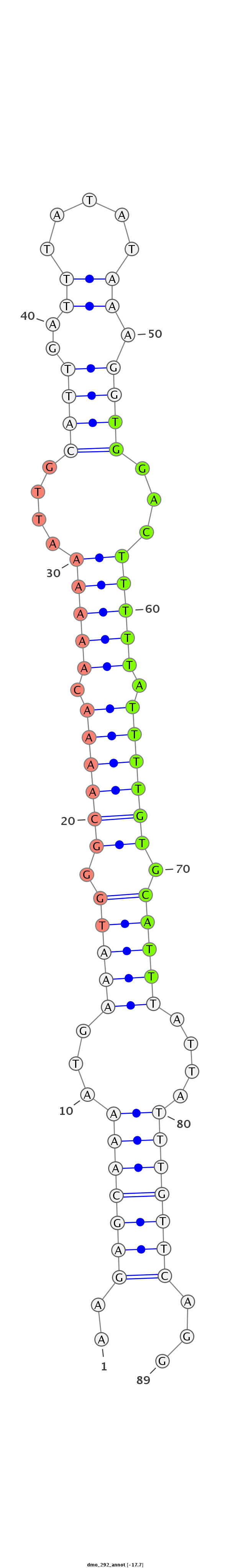
ID:dmo_292 |
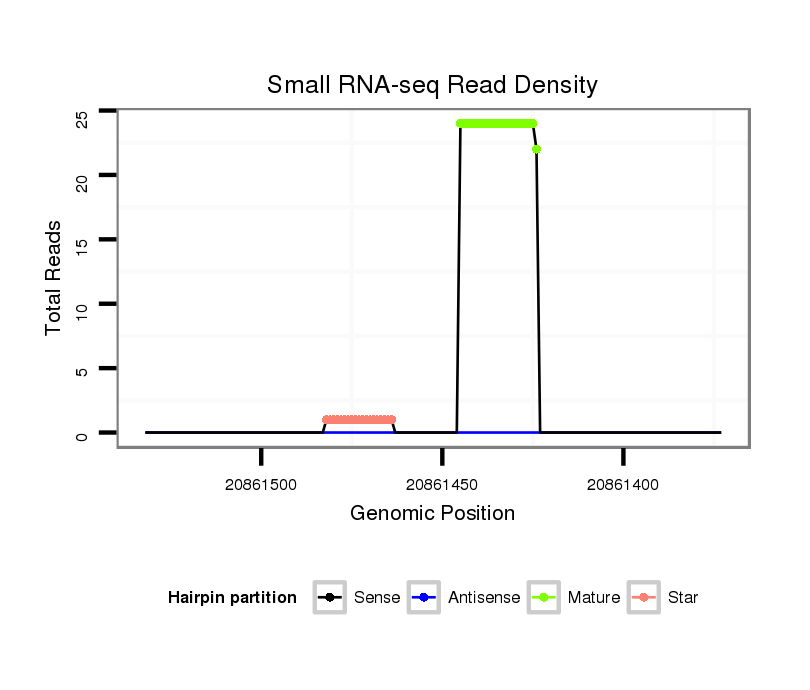
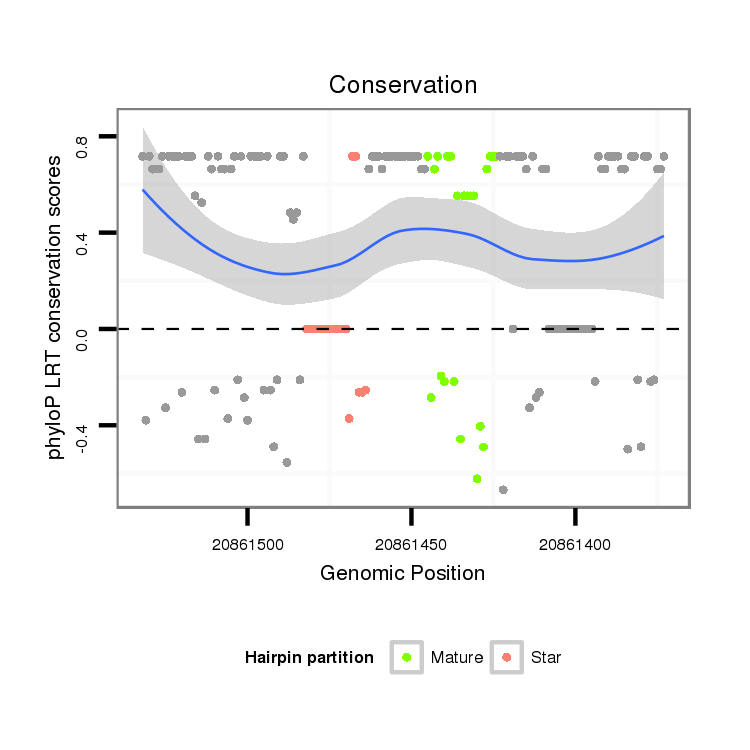
Coordinate:scaffold_6496:20861423-20861482 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -17.9 | -17.7 |
 |
 |
intron [Dmoj\GI18546-in]
No Repeatable elements found
|
ATTGGCATAAAATATATACATCGTGGTCAATGAAAAAGAGCAAAATGAAATGGGCAAAACAAAAAATTGCATTGATTTATATAAAGGTGGACTTTTTATTTTGTGCATTTATTATTTGTTCAGGGCGGCGAATGAAAATTGCAATTGCCATTGATTACGA
***********************************..(((((((...(((((.((((((.(((((....((((............))))...))))).)))))).)))))....)))))))...************************************ |
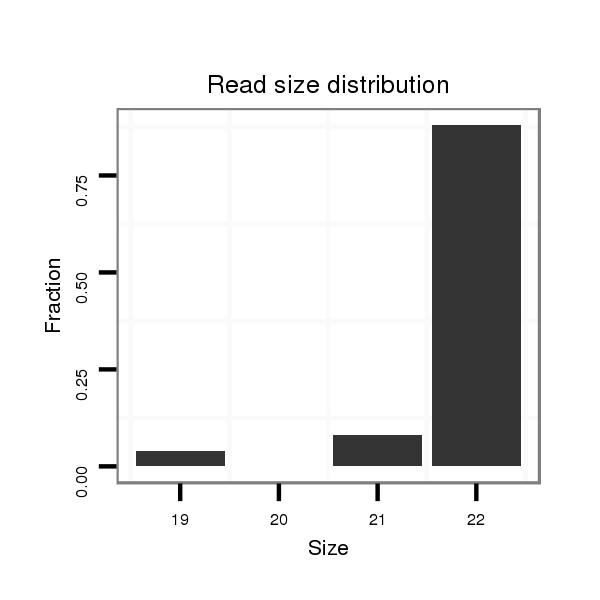
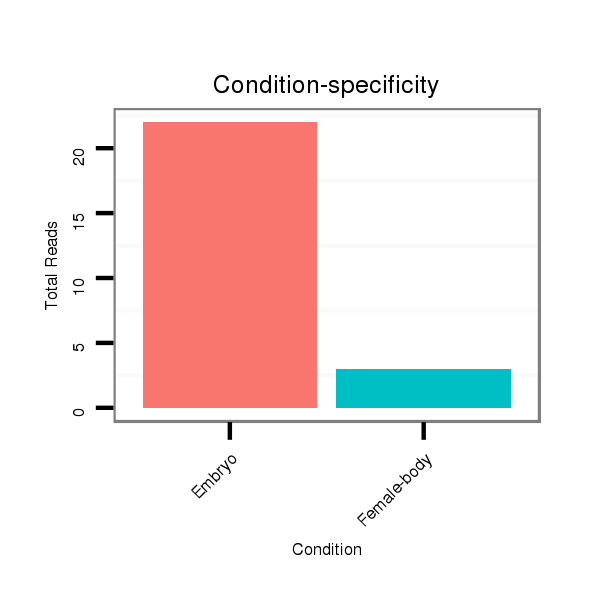
Read size | # Mismatch | Hit Count | Total Norm | Total | M060 embryo |
M046 female body |
V049 head |
V110 male body |
V056 head |
|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................TGGACTTTTTATTTTGTGCATT................................................... | 22 | 0 | 1 | 22.00 | 22 | 20 | 2 | 0 | 0 | 0 |
| .......................................................................................TGGACTTTTTATTTTGTGCATC................................................... | 22 | 1 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 | 0 |
| .......................................................................................TTGACTTTTTATTTTGTGCATT................................................... | 22 | 1 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 | 0 |
| .......................................................................................TGGACTTTTTGTTTTGTGCATT................................................... | 22 | 1 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 |
| .......................................................................................TGGACTTTTTATTTTGTGCAT.................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ................................................................................................TATTTTCTGCTTTTATTATTGGT......................................... | 23 | 3 | 19 | 1.79 | 34 | 0 | 30 | 0 | 3 | 1 |
| ..................................................TTGGCAAAACAAAAAATTGCATT....................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................TGGACTTTTTATATTGTGCATT................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................TGGTCTTTTTATTTTGTGCATT................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................TGCACTTTTTATTTTGTGCATT................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................TGGATTTTTTATTTTGTGCATT................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................TTATTTTCTGCTTTTATTATTGGT......................................... | 24 | 3 | 6 | 1.00 | 6 | 0 | 6 | 0 | 0 | 0 |
| ..................................................TGGGCAAAACAAAAAATTG........................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................TTCACTTTTTATTTTGTGCATT................................................... | 22 | 2 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 |
| .......................................................................................TTGACTTTTTATTTTGTGCATC................................................... | 22 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................ATTATTTTCTGCTTTTATTATTTGT......................................... | 25 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................TGGACGTTTTATTTTCTG....................................................... | 18 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 |
|
TAACCGTATTTTATATATGTAGCACCAGTTACTTTTTCTCGTTTTACTTTACCCGTTTTGTTTTTTAACGTAACTAAATATATTTCCACCTGAAAAATAAAACACGTAAATAATAAACAAGTCCCGCCGCTTACTTTTAACGTTAACGGTAACTAATGCT
************************************..(((((((...(((((.((((((.(((((....((((............))))...))))).)))))).)))))....)))))))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V110 male body |
M046 female body |
V056 head |
|---|---|---|---|---|---|---|---|---|
| ................................................................................................ATAAAAGACGAAAATAATAACCA......................................... | 23 | 3 | 19 | 0.21 | 4 | 1 | 3 | 0 |
| ........................................................................................................................GTCCTGGCGCTTACTTTTGA.................... | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 |
| .........................................................................................................................TCCTGGCGCTTACTTTTGA.................... | 19 | 3 | 17 | 0.12 | 2 | 0 | 1 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6496:20861373-20861532 - | dmo_292 | ATTGGCATAAAATATATACATCGTGGTCAATGAAAA-AGAGCAAAATGAAATGGGCAAAACAAAAAATTGCATTGATTTATATAAAGGTGGACTTTTTATTTTGTGCAT----TTATTATTTGT----TCAGGGCGGCGAATGAAAATTGCAATTGCCATTGATTACGA |
| droVir3 | scaffold_12875:4195078-4195206 + | ATTGGCAGAAAAAATA----TCATGGCCAATAAAAAAAAAACAAATTGAAA-------------GAAAAACATTGATTTATATAAAGGTAGACTTTT--------TCATGAAATTCTT-TTTGGGGTTTTTGG--------------CTGCAATTGCAATTGATTACGA | |
| droGri2 | scaffold_15112:2047928-2048060 - | AGTGGCAGAAAAAATATTCTTCATGGTCAGTGCAAAAAAAAGGAA----GA-------------AAAAAACATTGATTTATATAAAGGTGGATATTATTTTTTTCGCATCGACTTATT-TTTGG----TCTGG--------------CTGCAATTGCAATCCATAGCGA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/15/2015 at 03:04 PM