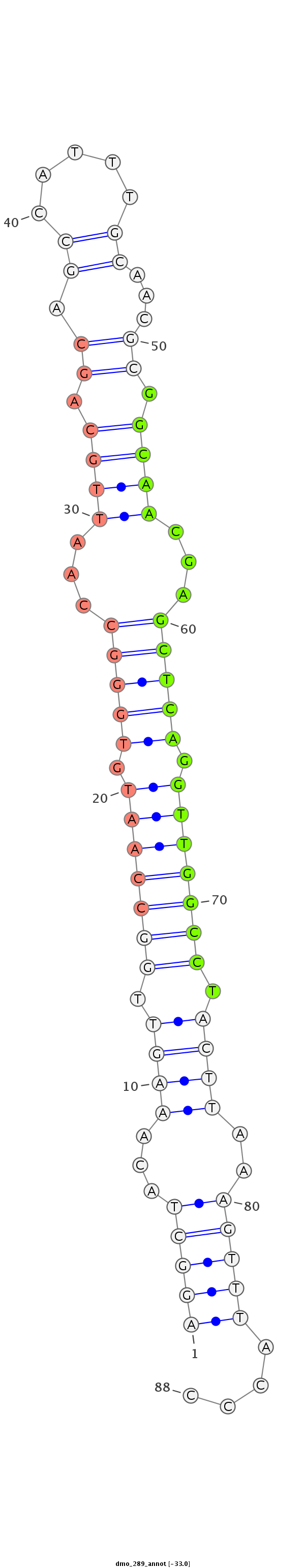
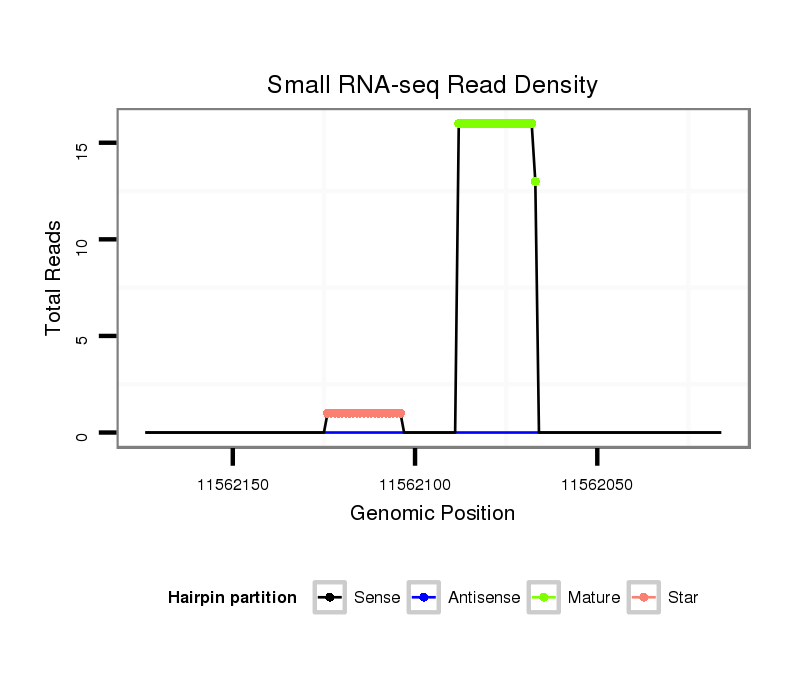
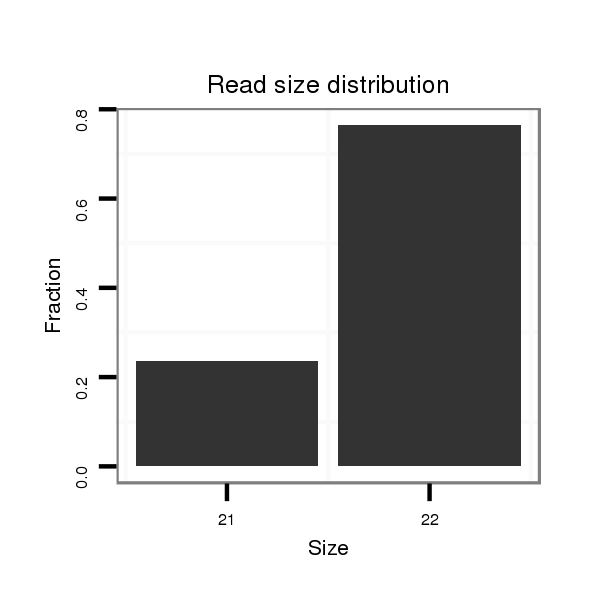
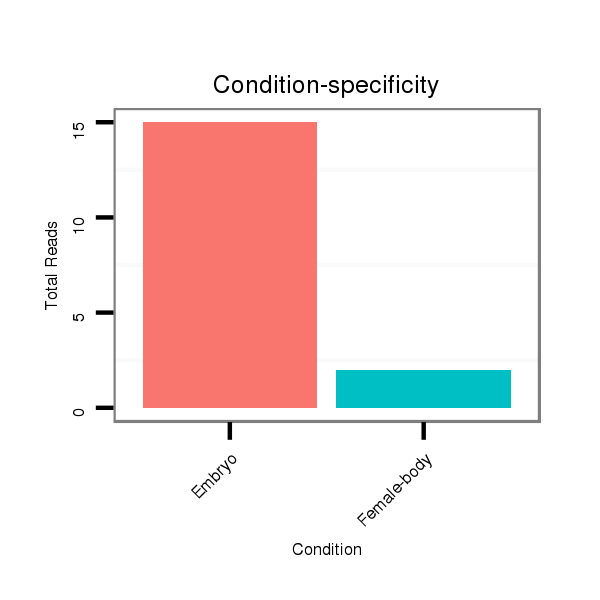
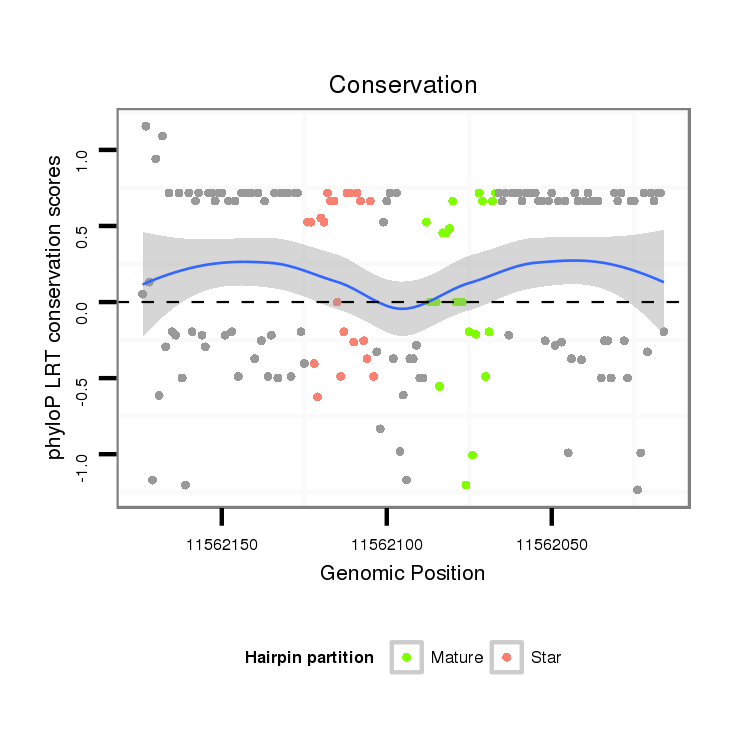
ID:dmo_289 |
Coordinate:scaffold_6540:11562066-11562124 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
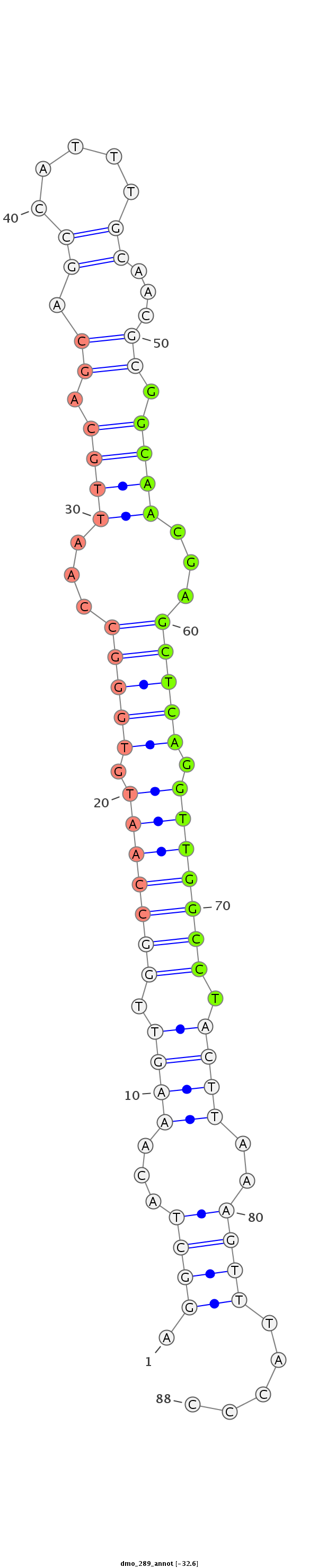
| -32.6 | -32.1 | -32.0 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CTCTGGCCAGTTGAAGCTACAACTTTGCCCAATAAAGGCTACAAAGTTGGCCAATGTGGGCCAATTGCAGCAGCCATTTGCAACGCGGCAACGAGCTCAGGTTGGCCTACTTAAAGTTTACCCCAGCTCATAGACAGGCGCCGACTCGGCCAAAACTTG
***********************************(((((...((((.(((((((.(((((...((((.((.((.....))...)).))))...))))).))))))).))))..)))))....************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | M060 embryo |
M046 female body |
|---|---|---|---|---|---|---|---|
| ......................................................................................GGCAACGAGCTCAGGTTGGCCT................................................... | 22 | 0 | 1 | 13.00 | 13 | 12 | 1 |
| ......................................................................................GGCAACGAGCTCAGGTTGGCC.................................................... | 21 | 0 | 1 | 3.00 | 3 | 2 | 1 |
| ......................................................................................GGCAACGAGCTCAGGTTGGCCC................................................... | 22 | 1 | 1 | 3.00 | 3 | 3 | 0 |
| ......................................................................................GGCTACGAGCTCAGGTTGGCCT................................................... | 22 | 1 | 1 | 2.00 | 2 | 2 | 0 |
| ......................................................................................GTCAACGAGCTCAGGTTGGCCT................................................... | 22 | 1 | 1 | 2.00 | 2 | 2 | 0 |
| ......................................................................................TGCAACGAGCTCAGGTTGGCCT................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 |
| ..................................................CCAATGTGGGCCAATTGCAGC........................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ......................................................................................GTCAACGAGCTCAGGTTGGCCC................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 |
| .................................................GGCAATGTGGGCCAATTGCAGC........................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 |
|
GAGACCGGTCAACTTCGATGTTGAAACGGGTTATTTCCGATGTTTCAACCGGTTACACCCGGTTAACGTCGTCGGTAAACGTTGCGCCGTTGCTCGAGTCCAACCGGATGAATTTCAAATGGGGTCGAGTATCTGTCCGCGGCTGAGCCGGTTTTGAAC
************************************(((((...((((.(((((((.(((((...((((.((.((.....))...)).))))...))))).))))))).))))..)))))....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V110 male body |
V049 head |
|---|---|---|---|---|---|---|---|---|
| .............................................................................................................GATTTTCCAATGGGGTGG................................ | 18 | 3 | 20 | 1.10 | 22 | 22 | 0 | 0 |
| ..................................................GGTTACACCCGGTTAGCATA......................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 |
| ...........................................................................................................ATGATTTTCCAATGGGGTG................................. | 19 | 3 | 12 | 0.08 | 1 | 0 | 1 | 0 |
| ...........................................................................................GCTCAAGTCCTACCGCATG................................................. | 19 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 |
| ........TCAGCTTGGATGTTGAAAG.................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6540:11562016-11562174 - | dmo_289 | CTCTGGCCAGTTGAAGCTA---CAACTTTGCCCAATAAA-GGCTACAAAGTTG------GCCAATGTGGGCCAATTGCAGCA-----GCCATTTGCAACGC----------------GGCAACGAGCTCAGGTTGGCCTACTTA-AAGTTTACCCCAGCTCATAGACAG--GCGCCGACTCGGCCAAAACTTG |
| droVir3 | scaffold_12822:1550137-1550292 + | CTCCGTCCAGCTTGAGCTGGGCCAACTTTGCCCAATAGA-AGCCAAAAAGTTGCACCT--------TGG-CCAAATGTGGCCCA-----CACTCCTGGTTAGGG-----------------TCGAG---GGTTTGGCCTACTTAAAAGTTTACCTCAACACTCAGACAG--GCTTTTACTTTGCGTACACTTG | |
| droGri2 | scaffold_14624:2593474-2593650 + | CTCG--CAAACTTCAACTG---AAACTTAGTCGAATAAAAAGGCAAAAACTTAAACGTAACCGCTGTGG-GTAAATGTAGGCCAGAACCCATTACGAACTAAACTGGTACAGGCAGCG-------G---CAACTGCTCTACTAAGAAGTTTACCTCAGCACGTAGCCAGAAGCTTTTACTTTGCAGACACTTA | |
| droEre2 | scaffold_4690:6767740-6767746 + | ATTGGGC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/15/2015 at 03:03 PM