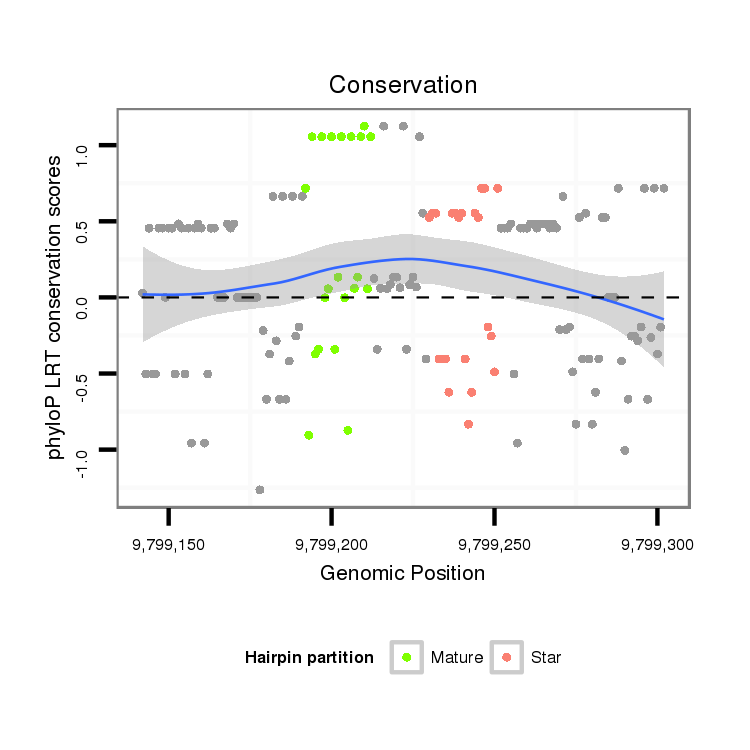
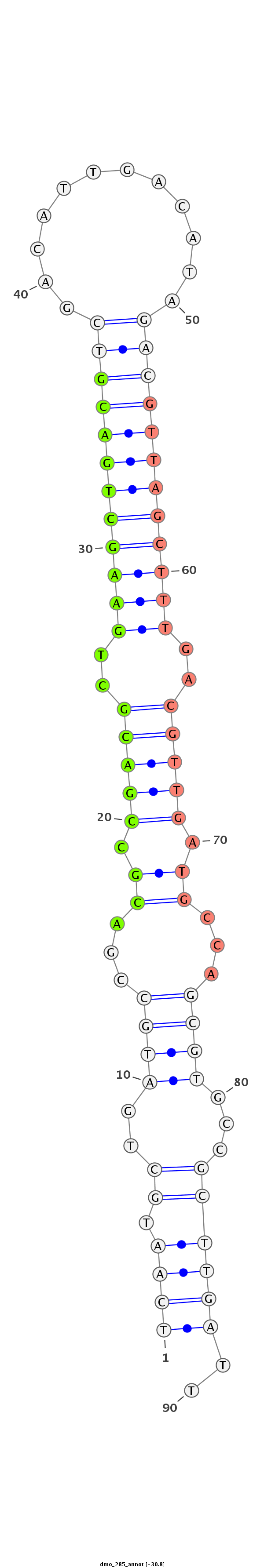
ID:dmo_285 |
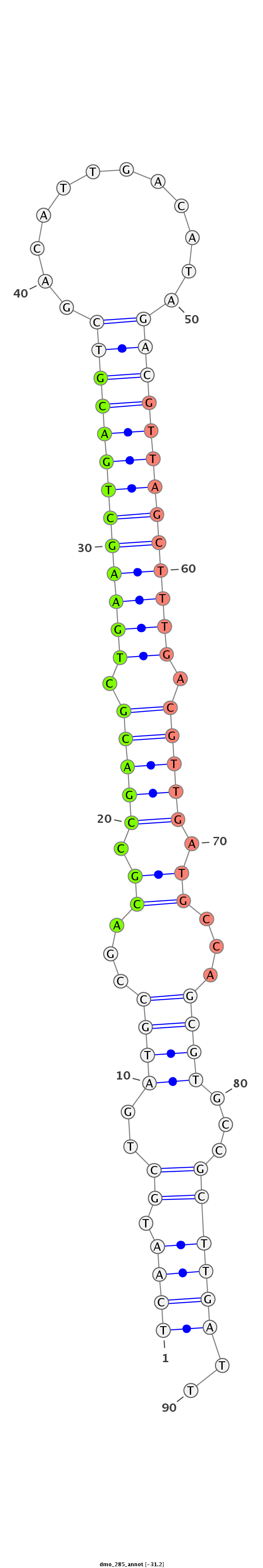
Coordinate:scaffold_6473:9799192-9799252 + |
Confidence:candidate |
Class:Canonical miRNA |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -30.8 | -30.4 | -30.3 |
 |
 |
 |
intergenic
No Repeatable elements found
|
AAGTGCGTGGTACAGCCACCAGCTGAACATGGAATTCAATGCTGATGCCGACGCCGACGCTGAAGCTGACGTCGACATTGACATAGACGTTAGCTTTGACGTTGATGCCAGCGTGCCGCTTGATTGACACTCGGCGTACTACCAGGAAGAGCGGAAAAACA
***********************************((((.((..((((...((.(((((.(((((((((((((............))))))))))))).))))).))...))))...))))))..************************************ |
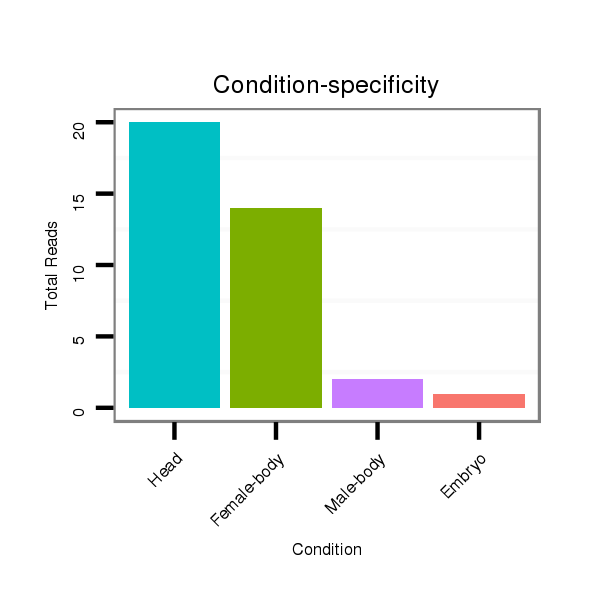
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V056 head |
V049 head |
V110 male body |
M060 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
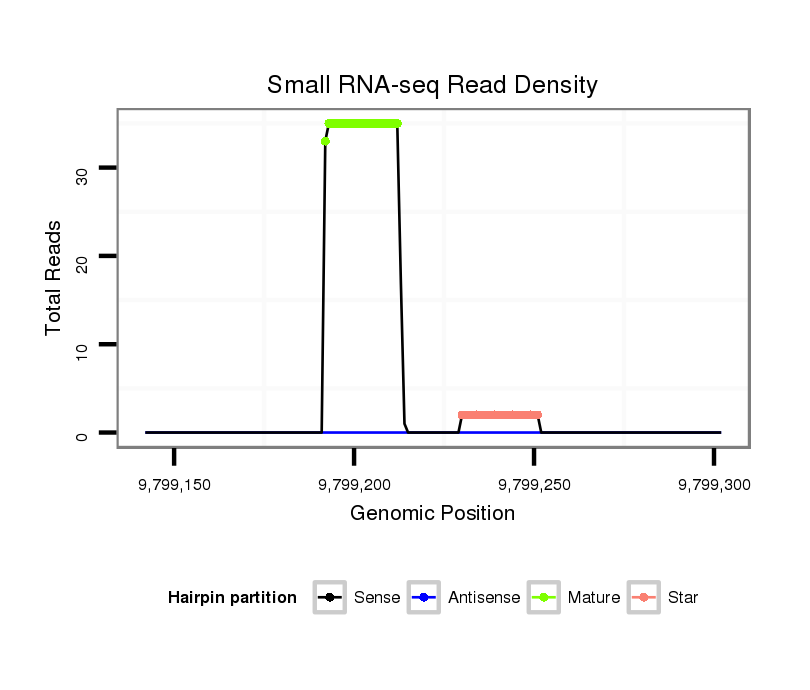
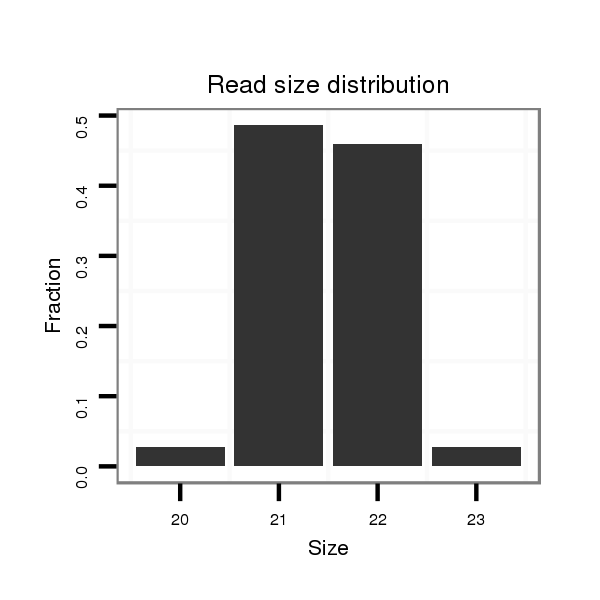
| ..................................................ACGCCGACGCTGAAGCTGACG.......................................................................................... | 21 | 0 | 1 | 17.00 | 17 | 1 | 10 | 5 | 1 | 0 |
| ..................................................ACGCCGACGCTGAAGCTGACGT......................................................................................... | 22 | 0 | 1 | 15.00 | 15 | 8 | 3 | 2 | 1 | 1 |
| ........................................................................................GTTAGCTTTGACGTTGATGCCA................................................... | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..................................................ACGCCGACGCTGAAGCTGACGTT........................................................................................ | 23 | 1 | 2 | 2.00 | 4 | 3 | 1 | 0 | 0 | 0 |
| ...................................................CGCCGACGCTGAAGCTGACG.......................................................................................... | 20 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...................................................CGCCGACGCTGAAGCTGACGT......................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ACGCCGACGCTGAAGCTGACGTC........................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ACGCCGACGCGGTAGCTGACG.......................................................................................... | 21 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................ACGCCGACGCTGAAGATGACG.......................................................................................... | 21 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................GGCCGACGCTGAAGCTGACG.......................................................................................... | 20 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................ACGCCGACGCTGAAGCCGACG.......................................................................................... | 21 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................ACGCCGACGCTGAAGCTGACGC......................................................................................... | 22 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................CCATTGACATAGACATCAGCT.................................................................. | 21 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ACGCCGACGCGGTAGCTGGCG.......................................................................................... | 21 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
|
TTCACGCACCATGTCGGTGGTCGACTTGTACCTTAAGTTACGACTACGGCTGCGGCTGCGACTTCGACTGCAGCTGTAACTGTATCTGCAATCGAAACTGCAACTACGGTCGCACGGCGAACTAACTGTGAGCCGCATGATGGTCCTTCTCGCCTTTTTGT
************************************((((.((..((((...((.(((((.(((((((((((((............))))))))))))).))))).))...))))...))))))..*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V110 male body |
|---|---|---|---|---|---|---|
| .................................................................................................CTGCAACTACGGTCACCCAG............................................ | 20 | 3 | 7 | 0.14 | 1 | 1 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droMoj3 | scaffold_6473:9799142-9799302 + | dmo_285 | candidate | AAGTGCGTGGTACAGCCACCAGCTGAACATGGAATTCAATGCTGATGCCGACGCCGACGCTGAAGCTGACGTCGACATTGACATAGACGTTAGCTTTGACGTTGATGCCAGCGTGCCGCTTGATTGACACTCG-----------GCGTACTACCAGGAAGAGCGGAAAAACA |
| droVir3 | scaffold_12970:11471999-11472125 - | AGGCACG-GGCACGGGCACGGGC---ACA-------GGCCGTGGCAGTCGAAGTTGCCGTTGCCGTTGACGTTAACGTTAATGTTG------------------ATGTCAGCGTAGCGCTTGATTGACACTCGCTCGTAGC----------------ATTCATAGACTAGCA | ||
| droGri2 | scaffold_15245:14940855-14940970 - | T-----------------------------------TGATGCTGATGTTGATGCTGATGTTGATGCTGATGTTGATGTTAATGTTGATGTTGATGTTGATCGTGATATGA------------------GCCTCATAATGACTCACCATGGGGCC---AAAAATGAAATAATA | ||
| droRho1 | scf7180000777921:6011-6046 - | A--------------------------------------------------TGTCGACGCCGACGCCGACGACGACGCCAACGCTG-------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
Generated: 10/20/2015 at 06:50 PM