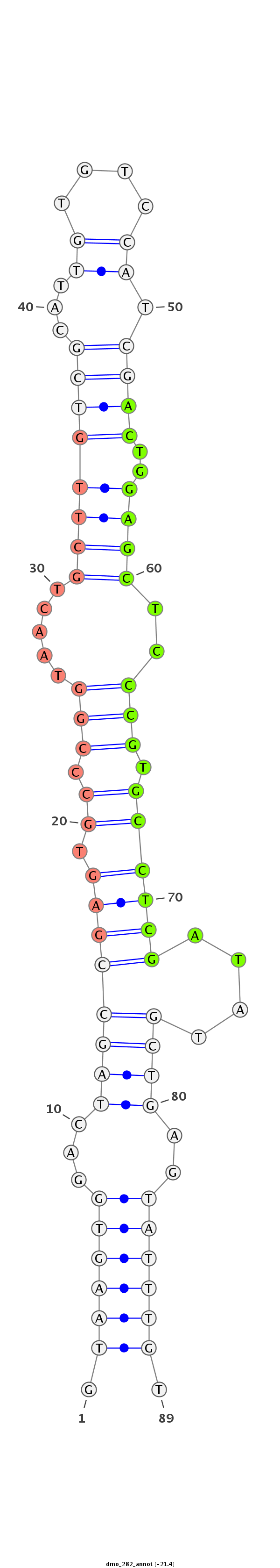
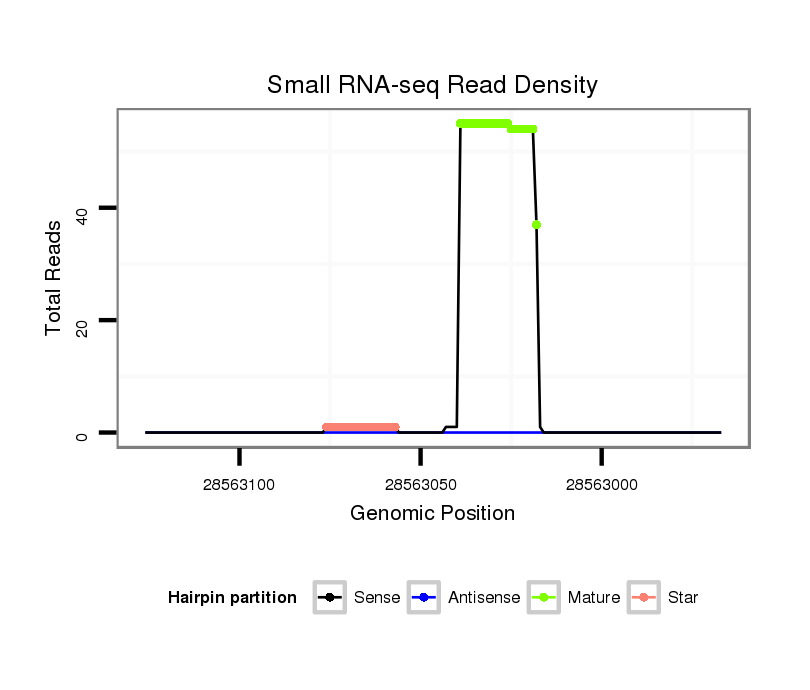
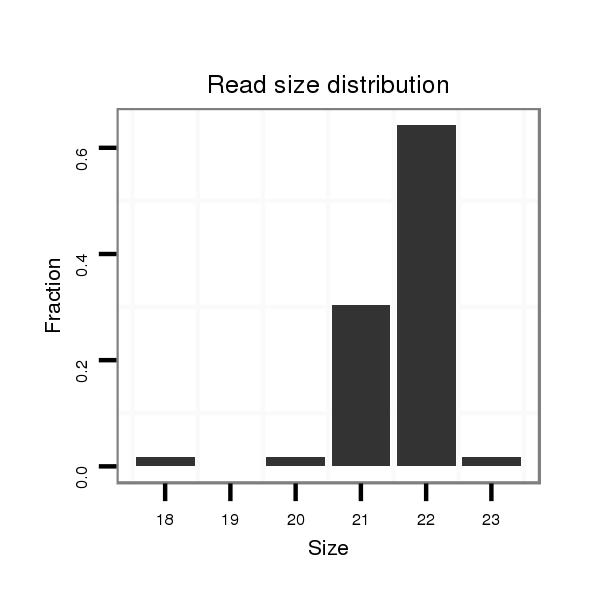


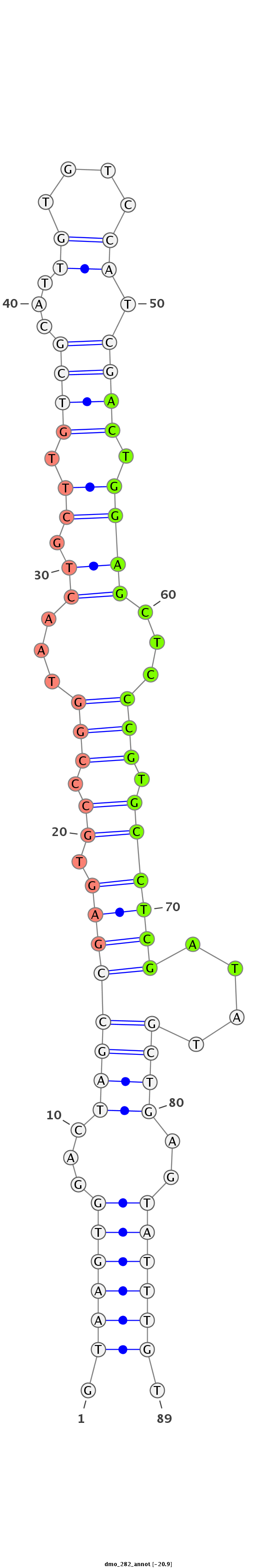
ID:dmo_282 |
Coordinate:scaffold_6540:28563017-28563076 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
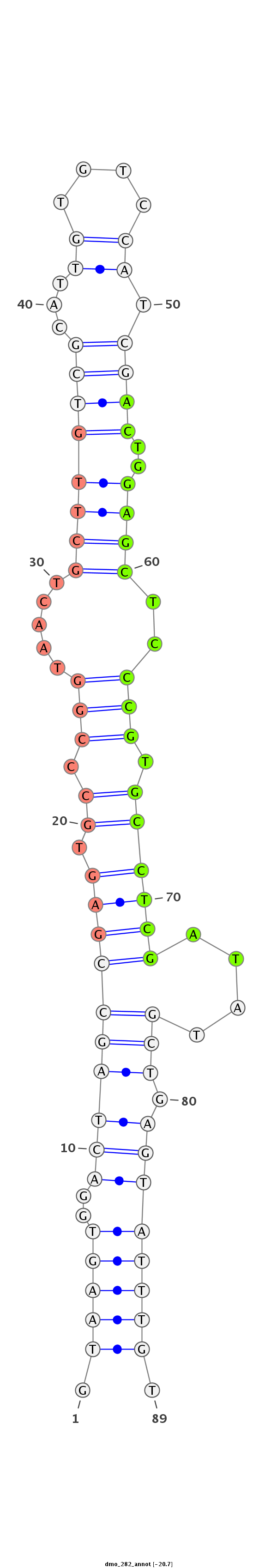
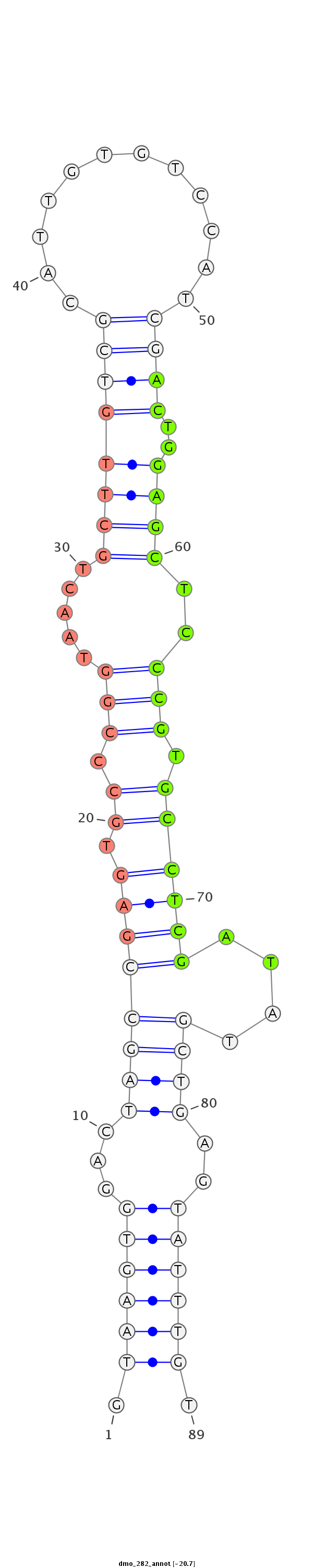
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -20.9 | -20.7 | -20.7 |
 |
 |
 |
intergenic
No Repeatable elements found
|
GTGCGCTTTGACCTAGAAGCCATTCTGCAAGACGGGTAAGTGGACTAGCCGAGTGCCCGGTAACTGCTTGTCGCATTGTGTCCATCGACTGGAGCTCCCGTGCCTCGATATGCTGAGTATTTGTCGTGCTGCATGAAGACACAGATGAACGCCATCCAGC
***********************************.((((((...((((((((.((.(((.....((((((((...((....)).))))..))))..))).))))))....))))..)))))).************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | M060 embryo |
V041 embryo |
M046 female body |
V056 head |
V110 male body |
V049 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................ACTGGAGCTCCCGTGCCTCGAT................................................... | 22 | 0 | 1 | 36.00 | 36 | 18 | 11 | 3 | 1 | 2 | 1 |
| .......................................................................................ACTGGAGCTCCCGTGCCTCGA.................................................... | 21 | 0 | 1 | 17.00 | 17 | 13 | 1 | 2 | 1 | 0 | 0 |
| .......................................................................................ACTGGAGCTCCCGTGCCTCGAC................................................... | 22 | 1 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GAGTGCCCGGTAACTGCTTG.......................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................ACTGGAGCTCCCGTGCCTCGT.................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................ACTGGAGCTCCCGTGCCTCGATT.................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................ACGGTTAAGCGGACTGGCCGA............................................................................................................ | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................ATCGACTGGAGCTCCCGT........................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................ACTGGAGCTCCCGTGCCTCGCT................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................AATGGAGCTCCCGTGCCTCGA.................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................ACTGGAGCTCCCGTGCCTCGATA.................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................ACCGGAGCTCCCGTGCCTCGAC................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TGTGCCCGGTAACTGCTTGTCG....................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................ACTGGAGCTCCCCTGCCTCGAT................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................ACCGGAGCTCCCGTGCCTCGA.................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................ACTGGAGCTCCCGCGCCTCGAT................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................ACTGGAGCTCCCGTGCCTCGTT................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................AGTGGAGCTCCCGTGCCTCGA.................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................ACTGGAGCTCCCGTGCCCCGAT................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................ACGGTTAAGCGGACTGGCC.............................................................................................................. | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................TTTTGTCAATGGACTGGAG.................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
|
CACGCGAAACTGGATCTTCGGTAAGACGTTCTGCCCATTCACCTGATCGGCTCACGGGCCATTGACGAACAGCGTAACACAGGTAGCTGACCTCGAGGGCACGGAGCTATACGACTCATAAACAGCACGACGTACTTCTGTGTCTACTTGCGGTAGGTCG
************************************.((((((...((((((((.((.(((.....((((((((...((....)).))))..))))..))).))))))....))))..)))))).*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V056 head |
V049 head |
M046 female body |
|---|---|---|---|---|---|---|---|---|
| ......................................TCACCTGTTTGGCTCACGGGCC.................................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..........................................................................................................................................TGAGGCTACTTGCGGCAGG... | 19 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 |
| ..........................................................................................................................................TGTGCCAACTCGCGGTAGG... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 1 |
| ..........TGGTTCTTCGGTCGGACGTT.................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6540:28562967-28563126 - | dmo_282 | GTGCGCTTTGACCTAGAAG--CCATTCTGCAAGACGGGTAAGTGGACTAGCCGAGTG---------CCCGG--T------------------AACT-GCTTGTCGCATTGTGTCCAT------CGACTGGAGCTCCCGTGCCTCGATATGCTGAGTATTTGTCGTGCTGCATGAAGACACAGATGAACGCCATCCAGC |
| droVir3 | scaffold_13047:10429985-10430159 - | GTGCGCCTCCACCTGGAAAGT--GTCCTGCTCGACGGGTAAGTGAACTAA----G---------TGTTCGGGCTTATCGCCTGG----CTCAAGGC-GCTTATTGTAACCTGCTTATTGTTGCTGGTTGGCTCTTTGGTGC---GCTGCGGCGCATGTATGCCTGGATGCATAAAGACACAGATGAACGCTATCCAGC | |
| droGri2 | scaffold_14624:151674-151814 - | CTAGACT----CCTGGAA-----------------------TCTCATCAG-CGAGTAAGTTTTTAC-TTGTTCTTGTTATCGATGTGGCTGAAAGTAGCTGACG-----------GC------TGCTT--------CCTTA---TGTGCTCCACAAATTTGCTGTGATTTAGGAAGACACACGCAAAAGCTAACCACA | |
| droEle1 | scf7180000491008:522214-522368 - | GCCCGTTTGCACCTGGAGGAGCTGTTTTCC-----AGGTAAGCCAGCCCG-CTAGTGACCTTGTCCTTCGCCCTCGTCCTCGCT-------------------CTCATCCTCCTAAT------CGACG-GACCT--CCTGCTGCG---------ATACTTCTCCGCCTGCTGGAACACACAGAGAAACGCCAGCCAAC | |
| droRho1 | scf7180000779477:313992-314133 - | GCCCGTTTGCACCTGGAAGAGCTGTTTTCC-----AGGTAAGCCAATTCG-CGAGTGACCTTGTCC-------------TCATC-------------------CTCACCCTCCTAGT------CGAGT-CTTGT--CCTGCTGCG---------ATACTTCTCTGCCTGCTGGAACACACACAGAAACGACAGCCAAC | |
| droTak1 | scf7180000415630:226434-226552 + | GTCCGTTTGCACCTGGAGGAGCTGTGGCTC-----AGGTAAGCGAACC------------TTGGTC-------------GCCTA-------------------ACCATCCT-------------------TCTT--CCTGCCGCG---------AAACTTGTCGGCCTGCCAGAAAACACACAGAAAGGCCAACCAAC |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/15/2015 at 03:03 PM