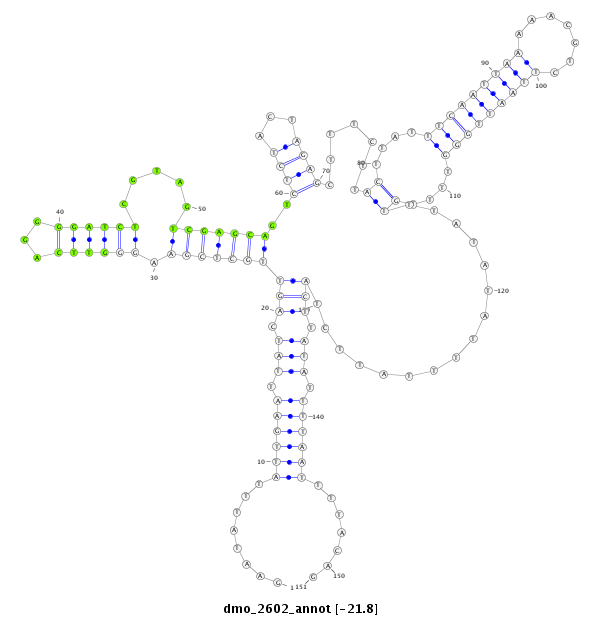
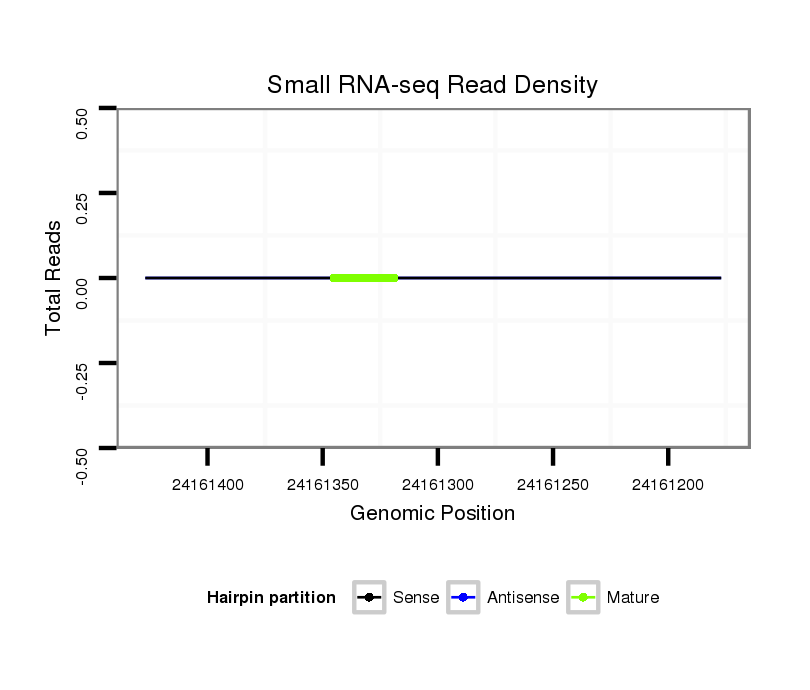
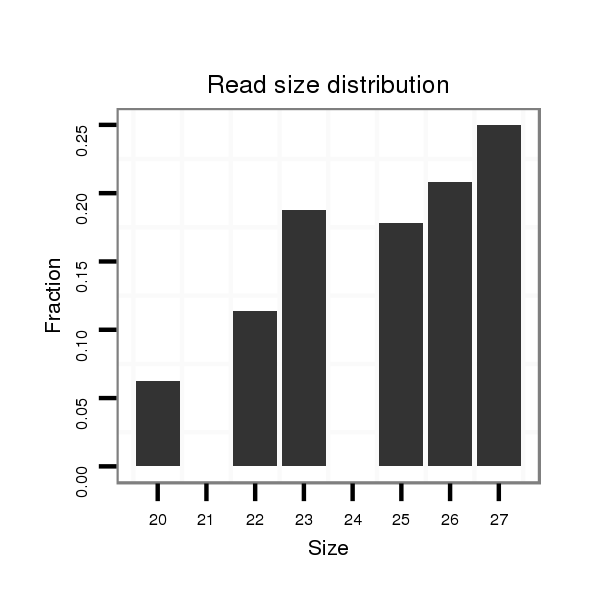
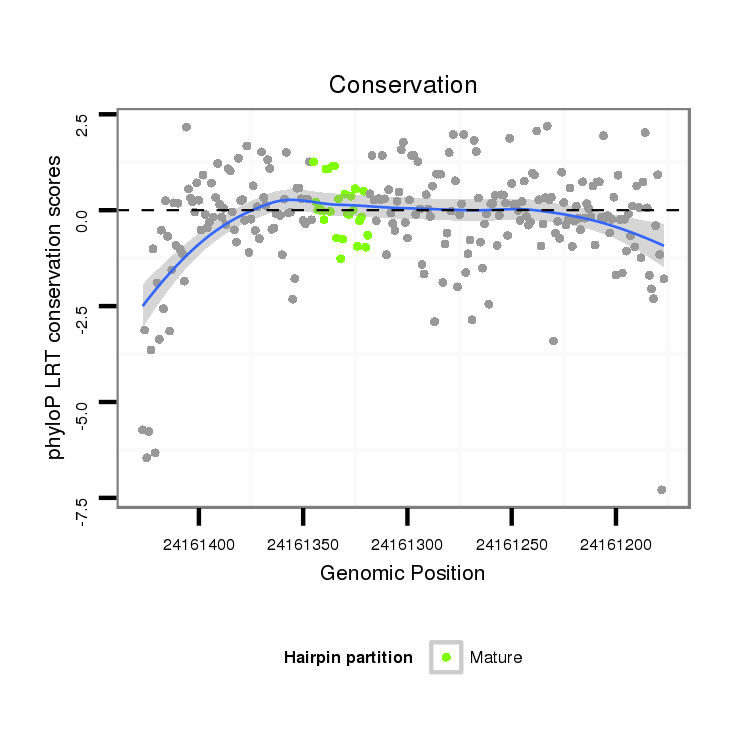
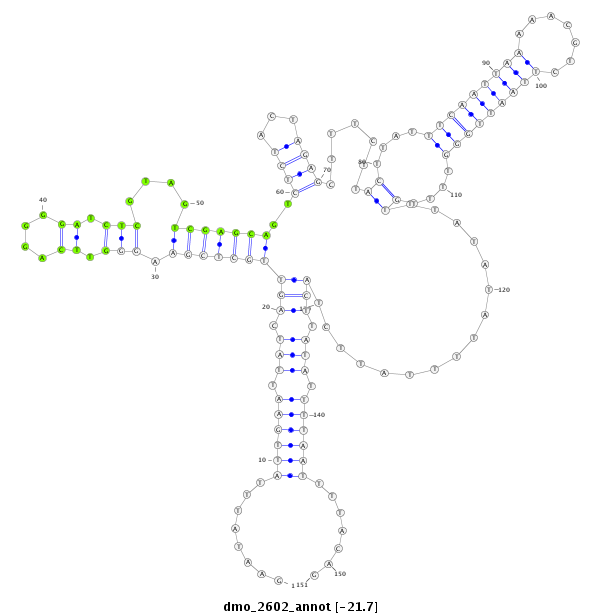
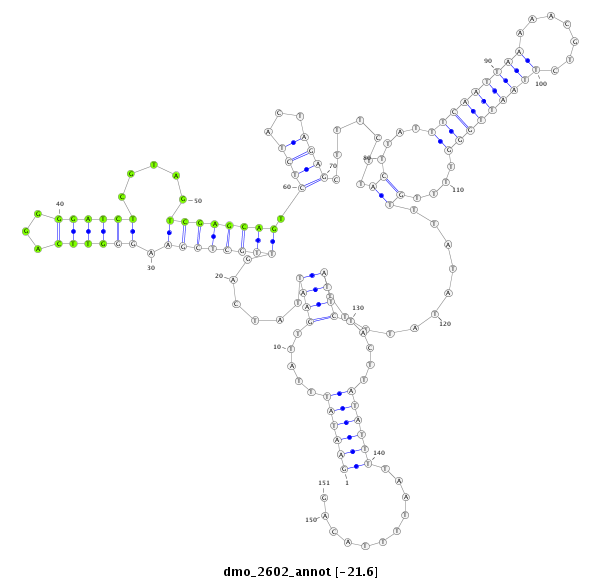
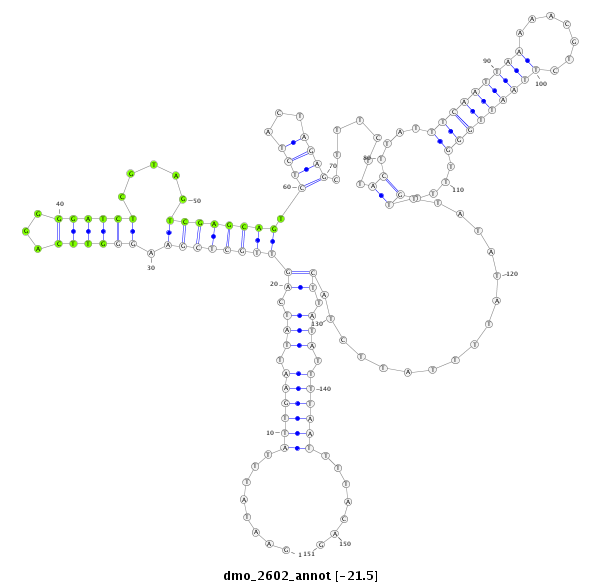
| AGCAATC-TT--TTTTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGTGACCTTT-----CTTACCGCAATGG------------------------------------------------------------ACACACTTTTAATTGGATT---------------TA-------------------------------------------------------------------------------------------------AACTT-------------CTTCATGGGTTTG----------------C---------------------------------------CTTA-------AATTTTGATTTCA-----------------------------------------------GA----TG----------- | Size | Hit Count | Total Norm | Total | M044
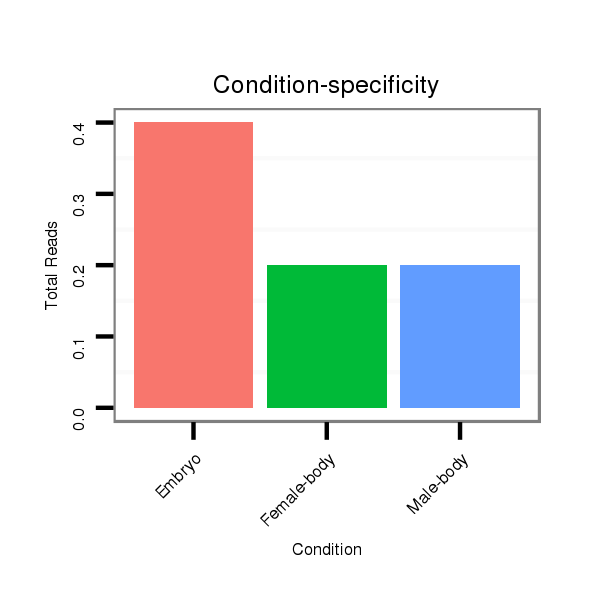
Female-body | M058
Embryo | V039
Embryo | V055
Head | V105
Male-body | V106
Head |
| .....................................................................................................................................................................................................TGACCTTT-----CTTACCGCAATGG------------------------------------------------------------ACACAC................................................................................................................................................................................................................................................................................................................ | 27 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................TGACCTTT-----CTTACCGCAATGG------------------------------------------------------------ACACACT............................................................................................................................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................TTGGATT---------------TA-------------------------------------------------------------------------------------------------AACTT-------------CTTCATGGGTTT................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................G------------------------------------------------------------ACACACTTTTAATTGGATT---------------TA-------------------------------------------------------------------------------------------------AACTT-------------C.............................................................................................................................................................. | 28 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................ATT---------------TA-------------------------------------------------------------------------------------------------AACTT-------------CTTCATGGGTTTG----------------C---------------------------------------CT........................................................................................ | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....ATC-TT--TTTTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGTGACCTTT-----CTTACC......................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............TG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGTGACCTTT-----CTTACCGCAATGG------------------------------------------------------------AC.................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................TAATTGGATT---------------TA-------------------------------------------------------------------------------------------------AACTT-------------CTTCATGGGT..................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........TT--TTTTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGTGACCTTT-----CTTACCGC....................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................TTACCGCAATGG------------------------------------------------------------ACACACTTTTAATTGG...................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................TAATTGGATT---------------TA-------------------------------------------------------------------------------------------------AACTT-------------CTTCATGGG...................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................TACCGCAATGG------------------------------------------------------------ACACACTTTTAATTG....................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................TT-----CTTACCGCAATGG------------------------------------------------------------ACACACTTTTAAT......................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................G------------------------------------------------------------ACACACTTTTAATTGGATT---------------TA-------------------------------------------------------------------------------------------------AAC.............................................................................................................................................................................. | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................TTGGATT---------------TA-------------------------------------------------------------------------------------------------AACTT-------------CTTCATGGGT..................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........TT--TTTTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGTGACCTTT-----CTTACCGCT...................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................TAATTGGATT---------------TA-------------------------------------------------------------------------------------------------AACTT-------------CTTCATGGGTT.................................................................................................................................................... | 28 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................ACCGCAATGG------------------------------------------------------------ACACACTTTTAATTGG...................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................CTT-------------CTTCATGGGTTTG----------------C---------------------------------------CTTA-------AATTT.......................................................................... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................G---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGTGACCTTT-----CTTACCGCAATGG------------------------------------------------------------ACA................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........T--TTTTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGTGACCTTT-----CTTACCGC....................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................TACCGCAATGG------------------------------------------------------------ACACACTTTTAATTGG...................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................TTAATTGGATT---------------TA-------------------------------------------------------------------------------------------------AACTT-------------CTTCATGG....................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................TGG------------------------------------------------------------ACACACTTTTAATTGGATT---------------TA-------------------------------------------------------------------------------------------------AACT............................................................................................................................................................................. | 28 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............TTTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGTGACCTTT-----CTTACCGCAATG................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................TTT-----CTTACCGCAATGG------------------------------------------------------------ACACACTTT............................................................................................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................GACCTTT-----CTTACCGCAATGG------------------------------------------------------------ACACACTT.............................................................................................................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................TTT-----CTTACCGCAATGG------------------------------------------------------------ACACACTTC............................................................................................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................TTAATTGGATT---------------TA-------------------------------------------------------------------------------------------------AACTT-------------CTTCATGGGT..................................................................................................................................................... | 28 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............TTTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGTGACCTTT-----CTTACCGCA...................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................TGG------------------------------------------------------------ACACACTTTTAATTGGATT---------------TA-------------------------------------------------------------------------------------------------AA............................................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............TTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGTGACCTTT-----CTTACCGCAATG................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............TG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGTGACCTTT-----CTTACCGCAATGG------------------------------------------------------------...................................................................................................................................................................................................................................................................................................................... | 26 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |