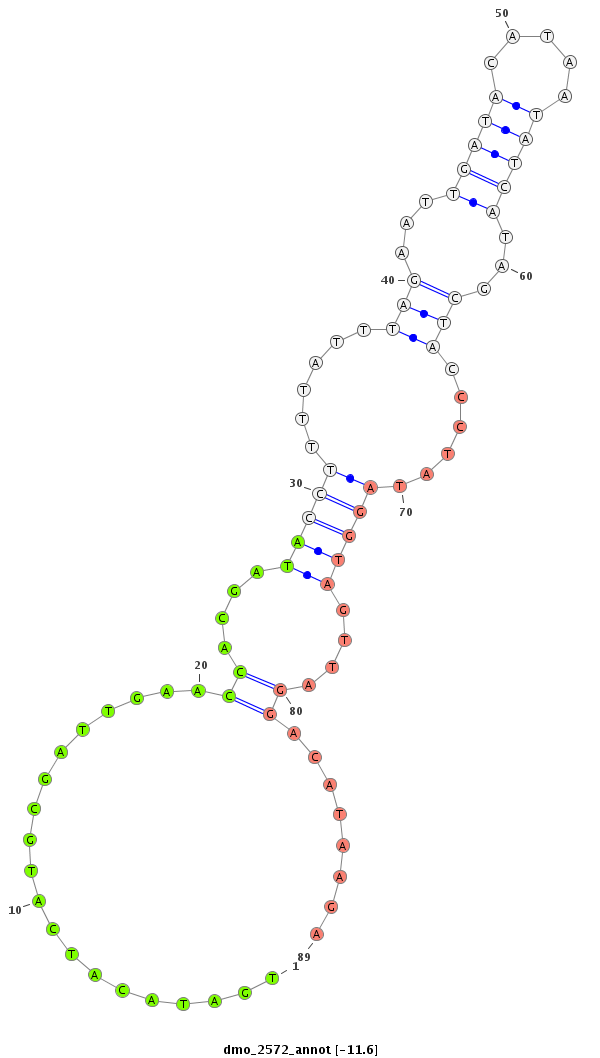



ID:dmo_2572 |
Coordinate:scaffold_6680:23203656-23203806 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -12.4 |
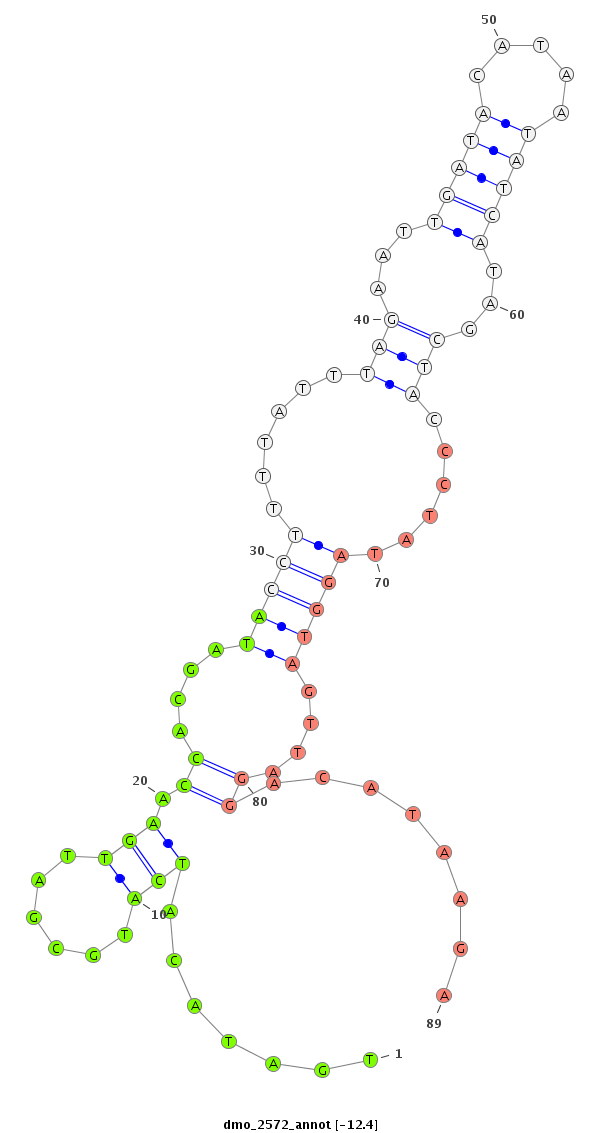 |
CDS [Dmoj\GI11335-cds]; exon [dmoj_GLEANR_11429:8]; intron [Dmoj\GI11335-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AATCAGCTTATAAGTCTTGATTTAAAACAAATGAATTCTCTTAAAGAAAGGTAAGACTTACTTTGTTTGTTTTTGTTTAATCCATATTATTTTTTGATACATCATGCGATTGAACCACGATACCTTTTATTTAGAATTGATACATAATATCATAGCTACCCTATAGGTAGTTAGGACATAAGATTTTTACCTTCTCAAAGGCTTGATGTTGGGGGATGATCGTAAATTTCTCAATACTCTTCAACAGTTCA **********************************************************************************************....................((....(((((......(((...(((((.....)))))...)))......)))))....))........******************************************************************** |
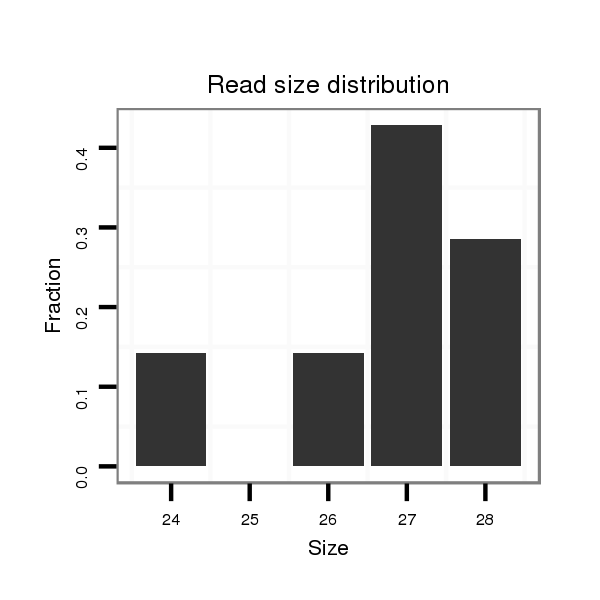
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V110 male body |
|---|---|---|---|---|---|---|---|
| ..............................................................................................TGATACATCATGCGATTGAACCACGATA................................................................................................................................. | 28 | 0 | 1 | 2.00 | 2 | 2 | 0 |
| ...............................................................................................................................................................................................TTCTCAAAGGCTTGATGTTGGGGGATGA................................ | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ...........................................................................TTTAATCCATATTATTTTTTGATACA...................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ...........................................................................................................................................................................................................................TCGTAAATTTCTCAATACTCTTCAACAG.... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ..........................................................................................................................................................................................TTACCTTCTCAAAGGCTTGATGTTGG....................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ........................................................................................................TGCGATTGAACCACGATACCTTTTATT........................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| .............................................................................................TTGATACATCATGCGATTGAACCACGA................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ...............................................................................................................................................................CCTATAGGTAGTTAGGACATAAGA.................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................................................................................................................TACCCTATAGGTAGTTAGGACATAAGA.................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................................................................................................................................................................GATGGTGGGGGCTGATCG............................. | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 |
| ..........................................................................................................CGATTGTACCACGATATATT............................................................................................................................. | 20 | 3 | 7 | 0.29 | 2 | 2 | 0 |
| ........................................................................................................................................................................AGTTAGGAGATAAGTTTCT................................................................ | 19 | 3 | 20 | 0.10 | 2 | 0 | 2 |
| .....................................................................................................................................................................GGAAGTTAGGAGATAAGTTT.................................................................. | 20 | 3 | 13 | 0.08 | 1 | 0 | 1 |
|
TTAGTCGAATATTCAGAACTAAATTTTGTTTACTTAAGAGAATTTCTTTCCATTCTGAATGAAACAAACAAAAACAAATTAGGTATAATAAAAAACTATGTAGTACGCTAACTTGGTGCTATGGAAAATAAATCTTAACTATGTATTATAGTATCGATGGGATATCCATCAATCCTGTATTCTAAAAATGGAAGAGTTTCCGAACTACAACCCCCTACTAGCATTTAAAGAGTTATGAGAAGTTGTCAAGT
********************************************************************....................((....(((((......(((...(((((.....)))))...)))......)))))....))........********************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V056 head |
V110 male body |
|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................CATCAATCCTGTATTCTAAAA................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .........................................................................................................CGCTAACATGGTGCTATATAA............................................................................................................................. | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ..........................................................................................................GCTAACATGGTGCTATATAA............................................................................................................................. | 20 | 3 | 7 | 0.29 | 2 | 1 | 0 | 1 |
| ..................................................................................................................................................................................................................CCCCTTAGGAGCATTTAAAG..................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 |
| ....................................................................................................................................CCTTAACTAGCTATTATAGT................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6680:23203606-23203856 - | dmo_2572 | AATCAGCTTATAAGTCTTGATTTAAAACAAATGAATTCTCTTAAAGAAAGGTAAG-----ACTTACTTTGTTTGTTTTTGTTTA--------------ATCCATATTATTTTTTGATACATCATGCGATTGAACCACGATACCTTTTATTTAGAATTGATACATAATATCATAGCTACCCTATAGGTAGTTAGGACATAAGATTTTTACCTTCTCAAAGGCTTGATGTTGGGGGATGATCGTAAATTTCTCAATACTCTTCAACAGTTCA |
| droVir3 | scaffold_13049:17913163-17913305 - | AACCAGCTAATAAGTATTGATTCAAAACAATTGACGTCTCTAAAAGAAAGGCGAGTAAAAATCAACTTTATTTGACTTTTATTATATATTTTGCTCGAATACCAATTATATTATTATAAAAT--------------------GTTT-----------------------------------------------AACATAAAACTATTAC------------------------------------------------------------A | |
| droGri2 | scaffold_10030:15832-15886 + | AATCAACTTATTAGCCTTGATTCAACACAGTTGACGTCACTTAAAGAAAGGTGAG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000780059:197799-197828 - | AATCAGCTTATCACAGTAGATGCACAACAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droBia1 | scf7180000302377:1761270-1761299 - | AATCAACTTATTACAGTGGATTCACAACAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
Generated: 05/18/2015 at 02:51 AM