
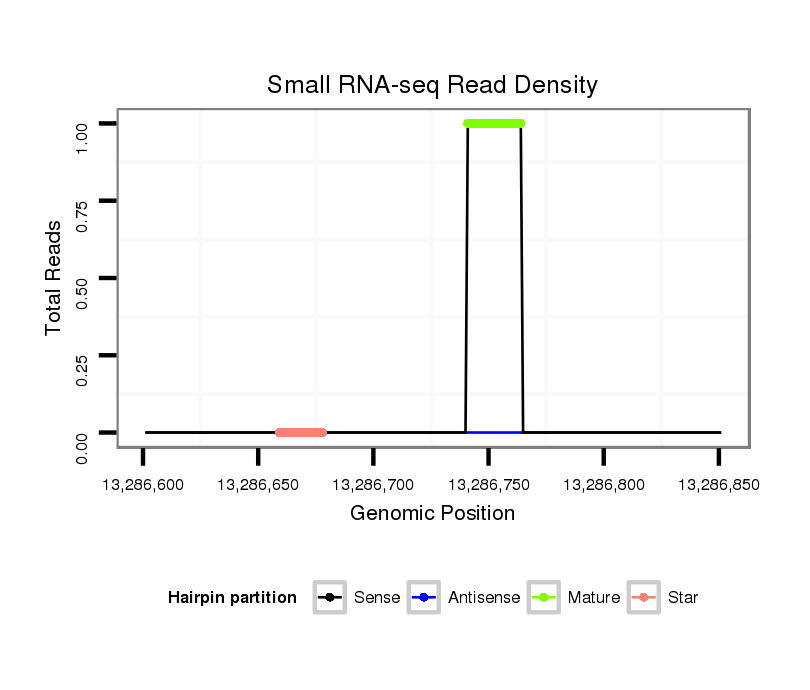
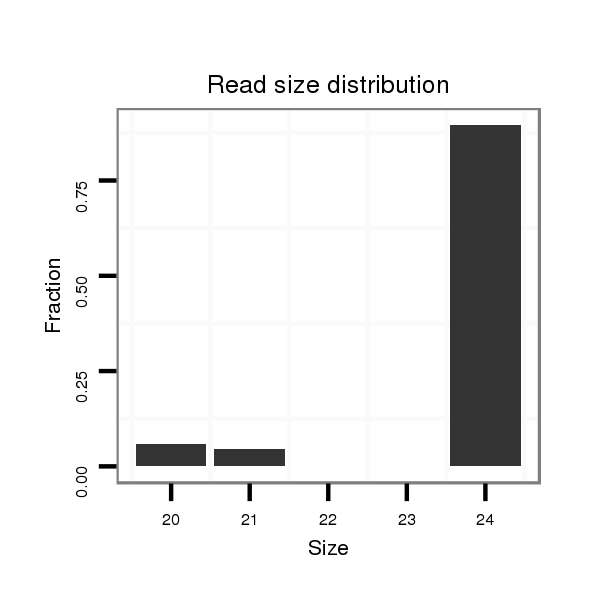
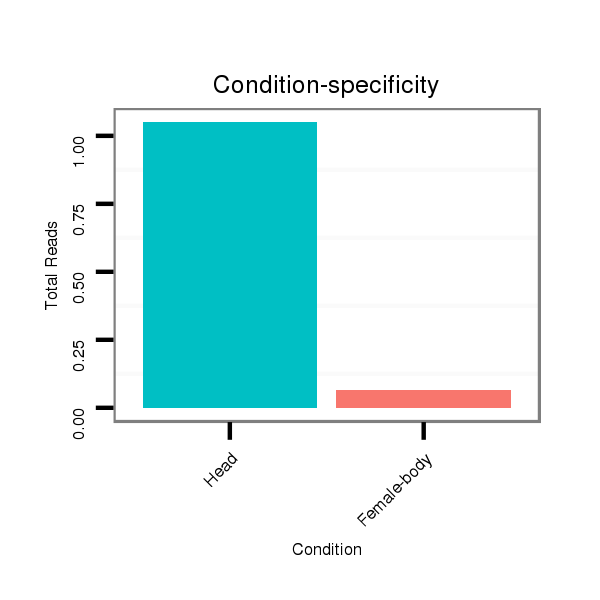
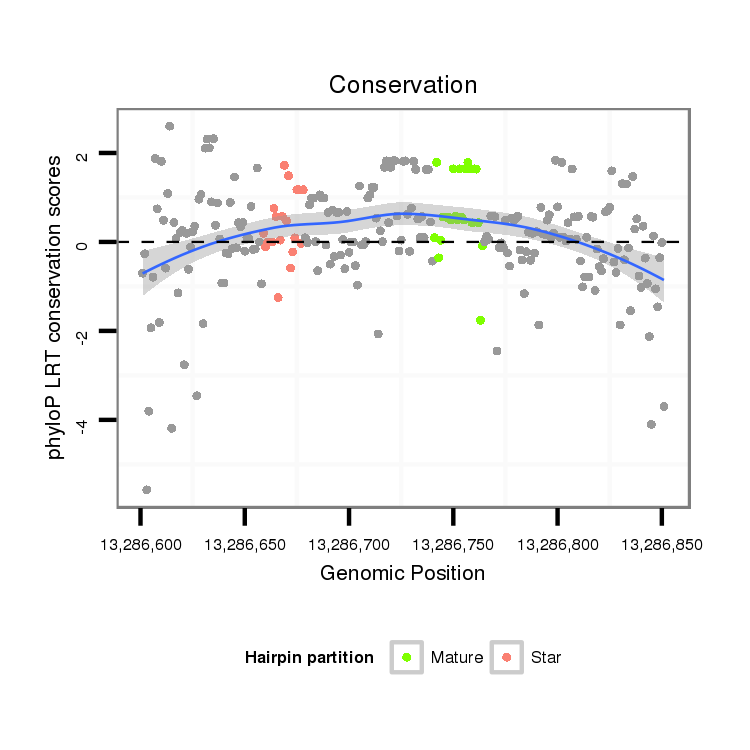
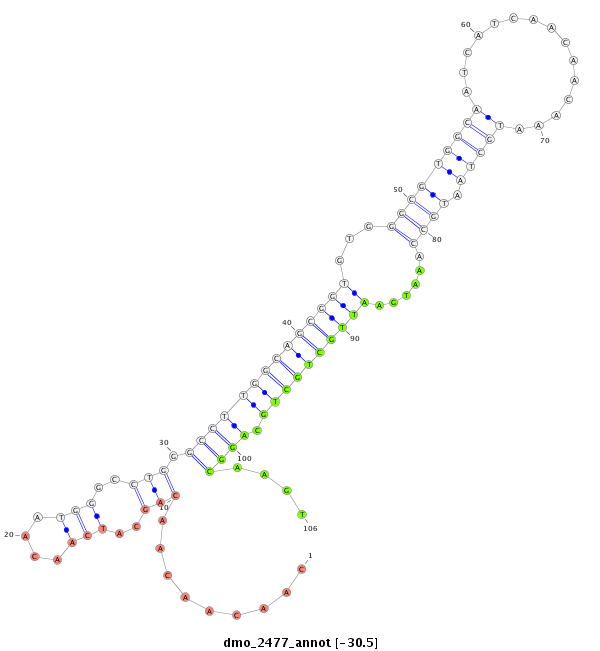
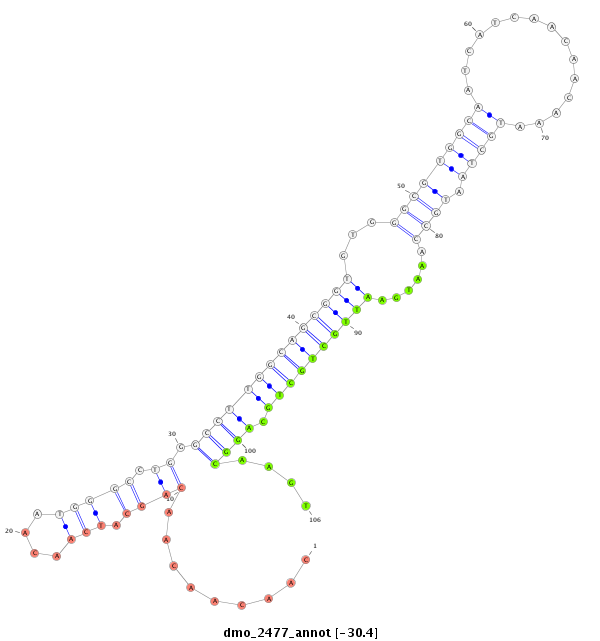
ID:dmo_2477 |
Coordinate:scaffold_6680:13286651-13286801 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -30.5 | -30.4 | -30.4 |
 |
 |
 |
CDS [Dmoj\GI13448-cds]; exon [dmoj_GLEANR_13373:21]; intron [Dmoj\GI13448-in]
| Name | Class | Family | Strand |
| TART_DV | LINE | Jockey | + |
| Gypsy8-I_Dpse-int | LTR | Gypsy | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GCACGACAGCAACATCGCTCACAGCAACAGCAGCAGAGATCGAACAACAAATTAACAACAACAACAACAGCATCAACAATGGGCCTGGGCCTTGGCAGCGGTGTGGGCGTGGCAATCATCAACAACAAATGCTAATGCCAAATGAATTGCTGCTGCAGGCAAGTCAATTAACAGCCGCCGACTGCAATCACAATTTAAGATATGTGCCGCCCCTGCCCACTGCCCCGCCCCCCATACAGGCAGCGGCCGGC **********************************************************.........(((..(((....)))..))).((((((((((((((...(((((((((...............)))).)))))......)))))))))).))))....*************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V049 head |
M060 embryo |
M046 female body |
V056 head |
|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................AATGAATTGCTGCTGCAGGCAAGT....................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................CCTGGGCCTGGACAGCGGT..................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ..........................................................CAACAACAACAGCATCAACA............................................................................................................................................................................. | 20 | 0 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 |
| ............................................................................................TGGCAGCGTTGTGAGCGGG............................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ......................................................ACAACAACAACAACAGCATCA................................................................................................................................................................................ | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
CGTGCTGTCGTTGTAGCGAGTGTCGTTGTCGTCGTCTCTAGCTTGTTGTTTAATTGTTGTTGTTGTTGTCGTAGTTGTTACCCGGACCCGGAACCGTCGCCACACCCGCACCGTTAGTAGTTGTTGTTTACGATTACGGTTTACTTAACGACGACGTCCGTTCAGTTAATTGTCGGCGGCTGACGTTAGTGTTAAATTCTATACACGGCGGGGACGGGTGACGGGGCGGGGGGTATGTCCGTCGCCGGCCG
***************************************************************************************.........(((..(((....)))..))).((((((((((((((...(((((((((...............)))).)))))......)))))))))).))))....********************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V110 male body |
M060 embryo |
V056 head |
V049 head |
V041 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................TCTACACGGCGGGGTCGG.................................. | 18 | 2 | 13 | 10.92 | 142 | 132 | 8 | 2 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................AATTGTCTACACGGCGGGGGC.................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................TCTACACGGCGGGGTCGGGT................................ | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................AAATTGTCTACACGGCGGG....................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................AATTGTCTACACGGCGGGGTCGG.................................. | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................ATTGTCTACACGGCGGGGTCGG.................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................CAAATTGTCTACACGGCGGGG...................................... | 21 | 3 | 2 | 1.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................TTGTCTACACGGCGGGGTCGG.................................. | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................CGGCGGGGTCGGTTGAGGGG.......................... | 20 | 3 | 20 | 0.45 | 9 | 1 | 8 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................CACGGCGGGGTCGGTTGAGGG........................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................TACACGGCGGGGTCGGTTGAG............................. | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................TCGTGTTAAATTCTTTACACGAC.......................................... | 23 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................ATTGTCTACACGGCGGGG...................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................GGGACGAGTGACGGGGCAGGT.................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................................................CTACACGGCGGGGACGGTT................................ | 19 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................TCGGCGGCGGACGCGAGTGTT.......................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................................................CACGGCGGGGTCGGTTGA.............................. | 18 | 2 | 13 | 0.23 | 3 | 2 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................AATTGTCTACACGGCGGGGTC.................................... | 21 | 3 | 9 | 0.22 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................ATTGTCTACACGGCGGGGTC.................................... | 20 | 3 | 15 | 0.20 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................TGTCTACACGGCGGGGTCGG.................................. | 20 | 3 | 16 | 0.19 | 3 | 2 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................AATTGTCTACACGGCGGGGT..................................... | 20 | 3 | 17 | 0.18 | 3 | 2 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................CTACACGGCGGGGTCGGG................................. | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................TTGTCTACACGGCGGGGTCG................................... | 20 | 3 | 12 | 0.17 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................TTTACACGGCGGGGTCGG.................................. | 18 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................GTCGTAGTTGTTACCAGGC..................................................................................................................................................................... | 19 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................CGGCGGGGTCGGTTGAGGGGG......................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................TCTATACACGGCGGGGTCA................................... | 19 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................TGTGGGTGGCTGACCTTAG.............................................................. | 19 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................GGGACGAGTGAGGGGGCGGGT.................... | 21 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................AAATTGTCTACACGGCGGGGT..................................... | 21 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................TCTACACGGCGGGGTCGCGT................................ | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................CACGGCGGGGTCGGTTGAGG............................ | 20 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................TCGTCTATATCTTGTTGGTT........................................................................................................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................AAATGTCTACACGGCGGGG...................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................................................................ACGTCCGATCAGGTAATG................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6680:13286601-13286851 + | dmo_2477 | GCAC-GACA--GCA---------------------------------------------------------------------------------A---------CATCGCTCACAGCAACAGCAGCAG-------------------------------------------------------------------------AGATCGAACAACAAATTAACAACAACAACA------------------------------ACAGCA------------------------------T--------------------------------------------------------------------------------------------------------------------------------------------------------CAACAATGGGCCTGGGCCTTGGCAGCGGT-----GT--------------------------------------GGGCGTGG------------CAATCATCAACAACAAATGCTAATGCCAA-----ATGAATTGCTGCTGCAGGCAA--------------------------------------------------------------------------GTCAATTAACAGCCGCC--GACTGC---------------------------------------------------------------AATC-ACAATTTAAGATATGTGCCGCCCCT-------------------------------------------------------------------------------------------------------------GCCCACTGCCCCG-C----C---------------------CCCC--------------ATACA-----------------------------------G------------------------GCAGCGG-CCGGC |
| droVir3 | scaffold_13049:14027990-14028258 - | GTGGCC---CCA--AAGCCG------------------------------C-GACA------------------------------------GCAG---------CAGCAGCAACGTCAG------------CAGCAGCAG------------------CA------ACA----------------------------GCAACGGCTGAGCAACAAATTAGCAACA------------------------------ACAACAACAGCC------------------------------ACG-----------------------------------------------------------------------------------------------------------------------------------------G------------CAACAATGGGCTTGGGCATGGGCG-----------G--------------------------------------GGGCGTGG------------CGATCATCAACAACAAATGCTAATGCCAA-----ATGAATTGCTGCTGCAGGCAA--------------------------------------------------------------------------GTCAATTAGCTGCCGCC--GACTGC---------------------------------------------------------------AATC-AGAATTTAAGATATGTACCGCCCAT-------------------------------------------------------------------------------------------------------------GCCAAGTGCCACG-C----C---TA-------TG-------------------------ACACA-----------------------------------A------------------------CCCCTGG-CTGGC | |
| droGri2 | scaffold_15110:16173418-16173689 + | GCAGCT---CCA--AAGCCA------------------------------C-GACA---------------------------------------G---------------CAACAACATCGTCAGCAG---CAG------CA------------GCAACAA---CA-------------------------------GCAGCG------CAACAAATTGGCAACA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAACAACAATC------------------GGCCTGGGCATGGGAG-----------T--------------------------------------GGGCGTGGGGGGTGGGGCAACAATCATCAACAACAAATGCTAATGCCAA-----ACGAATTGCTGCTGCAGGCAA--------------------------------------------------------------------------GTCAATTAACTGCTGCC--GACTGT---------------------------------------------------------------AATC-AGAATTTAAGATATGTGCCGCCAAT-------------------------------------------------------------------------------------------------------------GCCAAGTGCCACG-T----C----------GCTG-----C------------AAC-----------AGCAGCAACA-----------------------A------------------------TTGTTGG-ACGGA | |
| droWil2 | scf2_1100000004762:4626686-4626943 + | ACAC-AACA--ACA---------------------------------------------------------------------------------G---------CAACAATACCAGCAGCAGCAGCAG---CA----------------------------------------------------------------G-----------CAACAAATTAGCGATG------ACAACAG------------------------GAACA------------------------------ACA-----------------------------------------------------------------------------------------------------------------------------------------A------------CAACAAT-------------------------------------------------------------------------GG------------CTATCATCAACAACAAATGCTAATGCCAA-----ACGAATTGCTGCTGCAGGCAA--------------------------------------------------------------------------CACAATTAGCTTCCACCTCGAATAC---------------------------------------------------------------AGTTGGAAATTTAAGATATGTGCCGCCCATAGTACCGCCTAATGCTGCCCACTTGCATTCACACCCACATCCATTGG-----------------------------------------------------------------------------------------------------C------------AAC-----------AGCAGCAACA------------------------------------------------GCAGCAG-ATGGT | |
| dp5 | 2:468824-469042 + | CGGC-AGCA--ACG------------------------------------------GCA--C---------------------------------------------ACAGCAGCAGGGCCAA--GAAG---TA---------------------A------------------------------------------------------CTACCTATTAACAA------------------------------------------------TCCACAAGTACACCAATCACCAATCACCAATG--------------------AA--C-----------------------------------------AACCAACAACAACA---------------------------------------------ACAGCTACAG------------A------------------------------------------A---A---CAAACACCCAAAGAAAGACATTTG-------------------------------------------------------------------------------------------------AC-----------------------------------------------AATCTTCTTGT------------ACTATCCCCCAAATGTAT--------------GC--------------------------------------------------------CAAT------------------------------------------C-----------------------------------------GATTCTG------TATCTCTGTC---------------ACT-C----T---------------------ACCCGTTCA--------------------------------------------------------------------------GCAGGT-GTCGGT | |
| droPer2 | scaffold_9:1077018-1077319 - | GCTGCC---CCA--CC-------------------------------------------------------------------------------A---------AATCTGCGACAGTTGAATCAGCAG---CAA------CA------------GCA---ACGTCA-------------------------------GCAGCACGCCAGCAGCAAATTGGCAACA-------------------------------------------------------------GCA-------------------------------------------------------------------------------------------------ACACGGGCTTT---------------------------------------AGCC------ACATCAAGATCGGGAATGGC-------GCCA--GTG---------------CCAG----TGCCAGTGCCAGTGCAAGTGCCAGTGG------------AGAACATCAAGCACAATTGCTAATGCCCA-----ATGAATTGCTGCTGCAGGCAT--------------------------------------------------------------------------CCCGATTAGGTTCTTTC--GCCACC---------------------------------------------------------------GTCG-ACAATTTGAGATATGTACCGCCGCTGACTCCGGTCAAT-----------------------------------------------------------------------------------------------------GGTCCAC--------------------TGCGACGGCCCC-------------------T-----------------------------------G------------------------GCGGAT-GGCGGG | |
| droAna3 | scaffold_13340:11764751-11765013 - | ACACCA---GC--AGCA------------------------------------------------------------------------------A---------CAACAGCAACTCCAACACCAGCAG---CAA------CA------------CCAACAA---CACTA----------------------------TCAACA-----------TGCCAT--CCA---ACAACAGCAGC---------------------AAC------------------------------------------AGGCTA----------GCAAAA-TCTCATATCGTGGAATTTTTACCACCACAGGCAACGCG--------------------ATGAATGCAGCAGCCGCAGCCGCAGCGG-CGCAACAGCAGCAGCAACAAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCAGCAGCTTCCCTCCCCGCAGCTG--------------------------------------------------AGCGT--------------------TC----------------------------------TGC-------------------------------------------------------------------------------------CCGGCCAAATGTCG----CCGCC-----------------------------------------------CTC--------------GAACA-----------------------------------GTCT---------------GG----GCAACAG-CTGGG | |
| droBip1 | scf7180000396589:2062936-2063144 - | GCAACA---AC--AACA------------------------------------------------------------------------------G---------CAG---CTGCAGCAACAACAGCAG---TTG------CA---------ACAACA---ACAGCAGCA----------------------------GCAACA-------------------A------CA------A---------C------------A--------------------------------------------------------------------------------------------------------------------GCAGTTACA------------------------------------------GCAACAACAACCGATCATGCAA------C---------------------------AAGATCCCC-----------------------------------------------------AGCAGCAACAGCAGTCG-----------------------------------------------------------CC-----------------------------------------------AAG----------GCTGCC--GACCAC---------------------------------------------------------------TCCC-ACAGG--ACGACAAACGCCGCGTAC-------------------------------------------------------------------------------------------------------------CCCGCAGTCCACC-A----C----------GCCG-----CAACCCATCCAGTCG---------------------------------------------------------------------------C-CCCAAC | |
| droKik1 | scf7180000302510:446471-446710 - | CAGC-AGCC--ACA------------------------------------------TCTCACGCAGTTGCAGACGCCGCATAATCAAAACTACCAGCAA------CAGCAGCAAGCTCAG------------GC----------------------------------------------------------------------------CCAAAA-GTTGCCACT------GCAACAG------CAACAGCAGCAGCAGCAACAGCA------------------------------AACGCAGCAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTAACTACCAGCAACAACACAATTACACTACCCAGCAACAGC---------------------------------------------------------------AGTC-AGTCCCTGAGAAACCCGCCTCAGCACAAC---------------------------------------------------------------------------------------------------------AACAAGCCCCAC-------TTCCCACTGCCG--------T--------AAGCGA---------------------------------------------------------------------------T-GCCAGC | |
| droFic1 | scf7180000454105:1053569-1053804 - | GCAC-AGCA--GCA---------------------------------------------------------------------------------A---------CAGCAGCAACAGCAACAACAGCAG---CAA------CA------------ACAGCAACATC----------------------------------AGCA-------------------G------CAACAGCAGC---------------------AAC------------------------------------------AGGTGCGCCTGCTCACGCAAAAGACACA-----------------AACTGTACTCATTCCA----TTCAACAACA---ACA------------------------------------------ACAACAACAATC---------------------------------------ACATA---------A---ACAA----CGCTCCTGCCAAGACAACAG-------------------------------------------------------------------------------------------------CC-----------------------------------------------AAGAAATCCAG------------TCTGTCCGCCGGGAACAC--------------AA--------------------CCAC------------------------------------------------------------------------------C-----------------------------------------ACGACCA------TATTGCAGGC------------------------------------------------------AGCCA---------------------------------------------------------------------------A-GCCGGC | |
| droEle1 | scf7180000491199:1914995-1915210 - | GCCC-AACA--GCA---------------------------------------------------------------------------------G---------CAACAGCAACAGCAGCAGCAGCAA---CAA------CAGCAGCAACAGCAACA---ACAGCA---GGCAATGGCTGTGGGCGGCTCGGCGG---------------------------------------------------------------------------GCGGACAAGTGCAACAGCA-------------------------------------------------------------------------------------------------ACAAGTGCAGCAGGTGCAGCAGCAGCAAGTCCAGCAGCAGCAGCAGCAAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGTTACAA-------------------------------------------------------------------CATC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACCAAATCATA--G---------------------------------ACGCCTCGGGCAACATAACA--------------------------------A------------------CG----GCAACGA-CGAGC | |
| droRho1 | scf7180000779333:133900-134104 + | GCCA-CCCA--CCA---------------------------------------------------------------------------------GCAACATCAGCATT-TGAGCAGCAACAA---------CAA------CA------------ACA---A---CGGCA----------------------------GCAACA-------------------C------AC------A---------C------AGCCAA--CAG--AATACGCACAACAACAACAACAACACATTGCCAGTG--------------------GG--G-----------------------------------------GCACAACAGCAACA---------------------------------------------GCAGCAGCATC------------------------------------------------------AACATC-------------------------------------------AACAG------------------------------------------------------------------------CAGCGTGCAACATTG----------------------------------------------------------------------------------------CAACAT--------------------CCGC------------------------------------------------------------------------------C-----------------------------------------GCTGCCAAATAGCA-----------------------------------------------------------------------------------------------------------------A------------------CATCAAGCAGGC-TTC-GG | |
| droBia1 | scf7180000302113:1788567-1788803 - | GCGC-AGCA--GCA---------------------------------------------------------------------------------G---------CATCTGCAGCAGCATCAGCAGCAGCAGCAGCATCAG------------------------CAGCA----------------------------GCAACA-------------------T------CT------G---------CAGCAGCAGCAGCAGCAGCC------------------------------CCAGCAGCAA-----------------------------------------------------------------------------------------------------------------------------------------------------T---------------------------TGTTACGTC-----------------------------------------------------AGCAGCAACGGCAACTGCCCACGCAGG-----------------------------------------------------------------------------------CTGCCTACCAG-----------------------------------------------------------------------------------------------------------------------------------------------------CAACAGCAGCAACATC-AGGAGCAACAGCAACAACAGCAACAGCAACTGAGGCAAT-----------------------------CGTTTGCCCACA-ATGCCTTCCCACTGC-G-----------------CAGCGG---------------------------------------------------------------------------C-AACAGC | |
| droTak1 | scf7180000415809:748253-748477 + | CCTC-GGC--TG--GCAATC------------------------------AGGCTA---------------------------------------ACAA------CAATAACAATAGCAG------------CACCAACAACA------------ACAATAA---TA-------------------------------GCAACA-------------------A------CA---------------------ACAACAAT-------------------------AACAACGAGGAGCAAGTT--------------------A----------------------------------------------AGCAACACCAACA---------------------------------------------GCAGCAACAGC------------------------------------------------------AACAACAGCAGG---------CATCC------------------------------------------------------------------------------------------------------------AGCATAGCGGCCGCTGCAGCAGC--------------------------------------------------CGCCATGTACATGGATCCGATGCGCTACCAACATCCGCAC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CACCCGCATCCG--------C---------ATCCG---------------------------------------------------------------------------C-ACCAGC | |
| droEug1 | scf7180000409802:1796186-1796415 + | GCAGCA---GC--AACA------------------------------------------------------------------------------A---------CAT---CAGCAGCAGCAGCAGCAG---CAG------CA------------ACAACAACATC----------------------------------AACA-------------------C------CAGCAGCAGCAACAC---C------AGCAAT------------------------------CACAGTCCCAACAG-----------------------------------------------------------------------CAGCA---A------------------------------TC------------CCAGCAGCAA--------------------------------------------------------------------------------------------------------------------CTTC--ATGCTTTAGCCCAGCAACATATGTTGCAACTGCAGCC------------------------------------------------------G-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAACACCATC---AGC-----------AGCAGCAACACAATGCCACAAGTGAATGATACAGTCAGTACCTGGGCTGGAACT----ATGGCGC-CTGGC | |
| dm3 | chr3L:7442994-7443244 + | GCTGCG---GCA--GCTGCC------------------------------G-TCCA------------------------------------TCAGCAA------CAGCAGCAGGTGCAG------------CAGCAGCAG------------------CA------ACA----------------------------GCAACA-------------------C------CA------T---------CAGCAGCAACAGCAACAGCA------------------------------CCAGCAGCAGGTTG--CTGCCGTTGCGATG---------------------GCGGCCATGGCCAATTTGTTGCCGCAGCAGCAGTT---------------------------------------------GCTGCTGCATC------------------------------------------------------AGCAGAATCTGC---------TATCG------------------AATGCGGCGG------------------------------------------------------------------------TGGCC------------------ACAACGGTAACGG-------------------------------------------------------------------CGGC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCCAAGTTCCA--T---------------------------------TCACTGGTCGCAATACA-----------------------------------A------------------------ACAGGAG-CAGCA | |
| droSim2 | 3l:7308378-7308625 + | GCTGCG---GCA--GCCGCC------------------------------G-TCCA------------------------------------TCAG---------CAACAGCAGCAGGTG------------CAGCAGCAG------------------CA------ACA----------------------------GCAACA-------------------C------CA------T---------CAGCAGCAACAGCAGCAGCA------------------------------CCAGCAGCAGGTTG--CTGCCGTTGCGATG---------------------GCGGCCATGGCCAATTTGTTGCCGCAGCAGCAGTT---------------------------------------------GCTGCTGCATC------------------------------------------------------AGCAGAATCTGC---------TATCG------------------AATGCGGCGG------------------------------------------------------------------------TGGCC------------------ACAACGGTAACGG-------------------------------------------------------------------CGGC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCCGGGTTCCA--T---------------------------------TCACTGGTCGCAATACA-----------------------------------A------------------------ACAGGAG-CAGCA | |
| droSec2 | scaffold_2:2891270-2891470 - | GGCC-GCCA--CCG---------------------------------------------------------------------------------G---------CAACACCAACGGCAACAGCAGCAA---CAA------CG------------GCAACAA---CA-------------------------------GCAACG-------------------C------CGACGGCAGC----------------------------------------------AATACCACGGTTCCAGCGCC---------------------------------------------------------TTTG----GGCAGCAGCAGCAACA------------------------------------------GCAGCAGCAACC---------------------------------------ATGCT-------------AATGCTGC---------AAGTG------------------GAGGCAACAG------------------------------------------------------------------------TGGCCAGCGGCACGG----------------------------------------------------------------------------------------CGATGCCAATGCCAATGCCATTTCCGC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGCCT---------------------------------------------------------------------------C-CTCCGC | |
| droYak3 | X:6616096-6616344 + | ACACCA---AA--GACAACAATGGCCAAGATCGGCGTGACAACAGCGGCA------GGAACCACTGTTGCCGAGGCAGGAAAA------------------------------------------------------------------------ACA---ATATCA---CCCAAGGGGCTCAAGCAATCTGGCAACG-----------------CCGTAGCCATG------ACAACAG--------------------------------------------------------------------------------------------------------------------------------------------------CA------------------------------------------ACACCAACAATC-----------------------------------------------------------------------------------------------------------------------------------AC-----ACCAACAACAGCAACAGGCAAATCAAAATTGAACGAAACAGC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGGGCCAAAAATAAATAGCGGGCAGCCGGCCAAG--------T----C-----------------------------ACCTGAACAACAACA-----------------------------------G------------------------GCAACGG-CAGCC | |
| droEre2 | scaffold_4784:2897661-2897899 - | GGCC-GCCA--CCG---------------------------------------------------------------------------------G---------CAACACCAACGGCAACAGCAGCAACAGCTGCAGCAACA---------ACGGCAACAG---CA-------------------------------GCAACG-------------------C------CGACGGCAGC----------------------------------------------AATACCACGGTTCCAGCGCC---------------------------------------------------------TCTG----GGCAGCAGCA---ACA------------------------------------------GCAGCAGCAACC---------------------------------------ATGCT-------------AATGCTGC---------AAGTG------------------GTGGCAACAA------------------------------------------------------------------------TGGCCAGCGGCACGG----------------------------------------------------------------------------------------CGATGCCAATGCCAATGCCATTTCCGCCGCCTCCGCCGCAGTAGGAAATCCGGGT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A-----------------------------------A------------------------CCAGCAG-CC-GA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/18/2015 at 02:43 AM