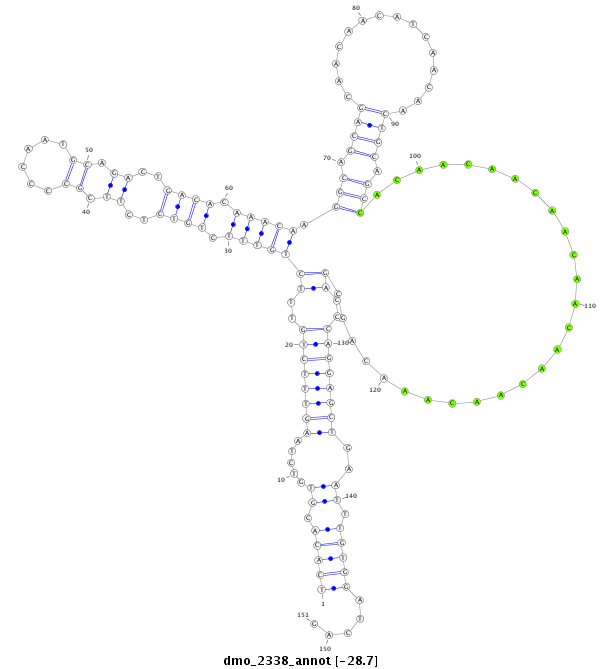
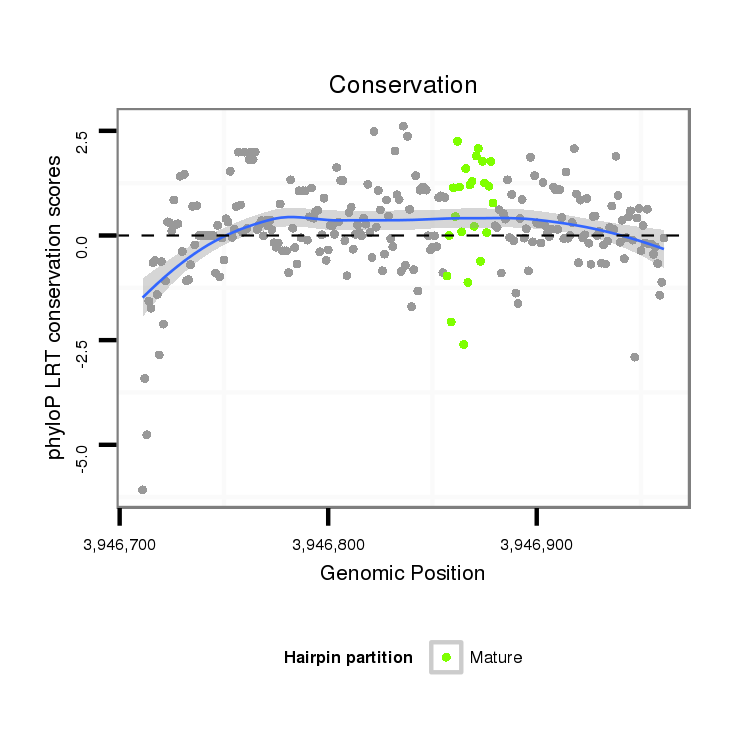
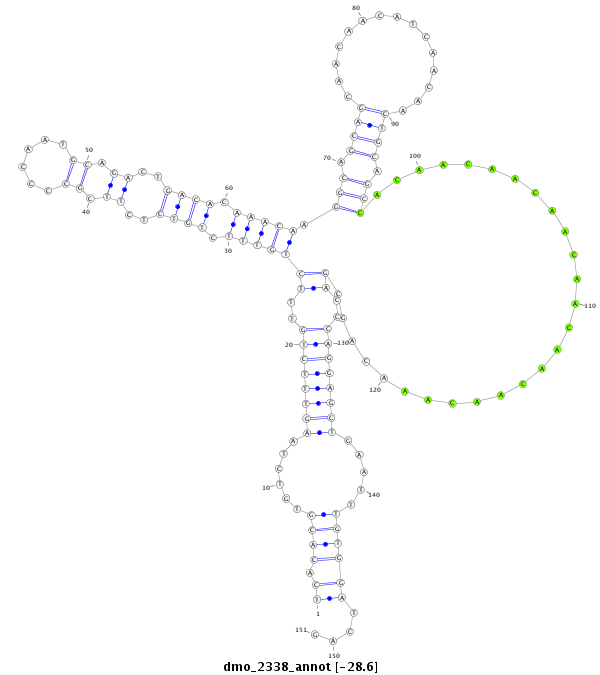
| ATCTTG------------------------------------------------------------------------------TAA--------------------------------------------------------------------------------------ATATTCT------------------------GTGCCTATTGCTAAACATTT----------------------------A-CTTC-----ATG--TTTTAG------AGCGGCTTTC--A-------------GCA---AC--AACAGAGACAGA-CCCCT----------------CAGCAGCAGCAGCAACAT---------------------------CAGCATC---------------AGCA------GCAGCAG---CA------GCAGCAGCA---G------CAACAT---CAGCAGCA-GCATCAGCAGCAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAACAT--------CAACATCA------------------GC | Size | Hit Count | Total Norm | Total | M026
Head | M043
Female-body | M056
Embryo | V046
Embryo | V052
Head | V058
Head | V120
Male-body | SRR1275488
Male larvae | SRR1275486
Male prepupae | SRR1275484
Male prepupae | GSM1528802
follicle cells |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................T--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAACAT--------CAAC........................ | 20 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GCAGCAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAACAT--------C........................... | 26 | 6 | 0.33 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAACAT--------C........................... | 22 | 10 | 0.30 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GCAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAACAT--------C........................... | 23 | 8 | 0.25 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAACAT--------C........................... | 20 | 12 | 0.25 | 3 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GCATCAGCAGCAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------............................................... | 20 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGCAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAACAT--------C........................... | 24 | 8 | 0.25 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGCAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAAC...................................... | 21 | 20 | 0.20 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAACAT--------C........................... | 21 | 11 | 0.18 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAACAT--------............................ | 20 | 13 | 0.15 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GCAGCAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAACAT--------............................ | 25 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAGCAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAACAT--------............................ | 24 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAACAT--------C........................... | 19 | 17 | 0.12 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAACA..................................... | 20 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGCAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAACAT--------............................ | 23 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................................................CA---G------CAACAT---CAGCAGCA-GC.................................................................................................................................................................................................. | 19 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAGCAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGC......................................... | 19 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAACAT--------............................ | 21 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GCAGCAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAAC...................................... | 23 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................GCA---G------CAACAT---CAGCAGCA-GC.................................................................................................................................................................................................. | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAAC...................................... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GCAGCAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGC......................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................ATC---------------AGCA------GCAGCAG---CA------GCA...................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................GCAGCAGCA---G------CAACAT---CAGCAGCA-.................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GCAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAACA..................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAGCAACAT--------------------------------------------------------------------------------------------------------------------------CAGC-------AGCAGCAA....................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................ATC---------------AGCA------GCAGCAG---CA------GCAGCAGCA---G------CA.................................................................................................................................................................................................................... | 28 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |