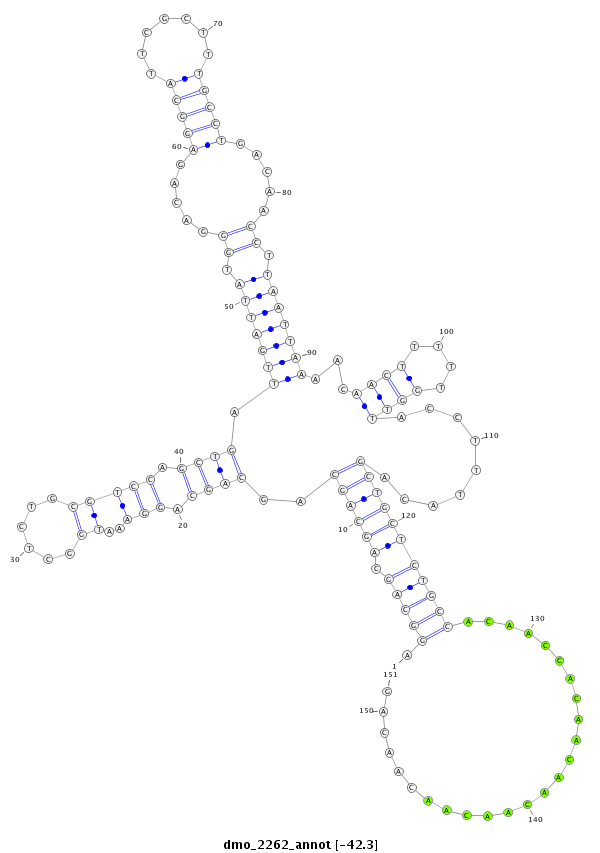
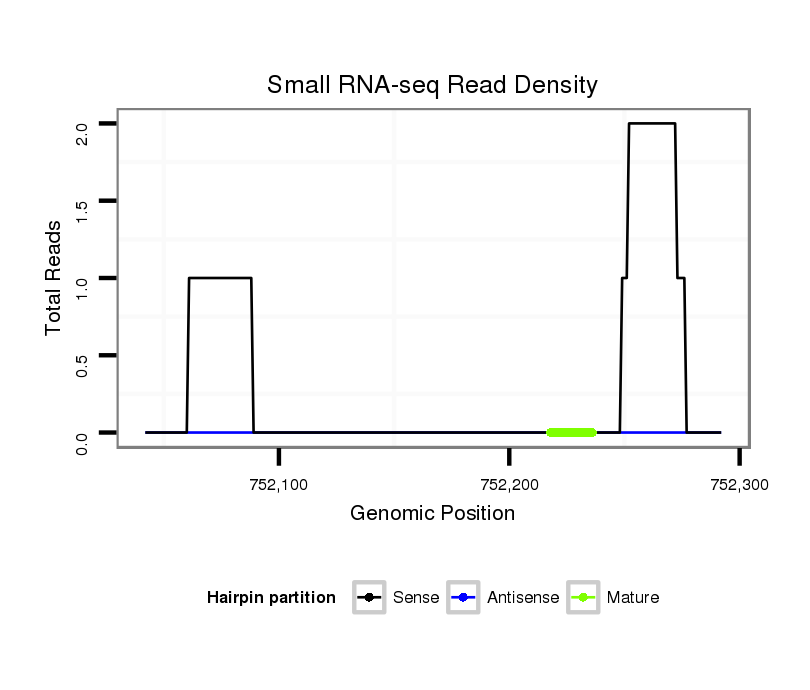
| droMoj3 |
scaffold_6654:752042-752292 + |
dmo_2262 |
GAT---------ATATTGGTGGCAGGCA---CATTGTAATAGAGCCAT-GCAAC----ACGAAGCAAAGGCAGCAGCAGCAG---------------CAGCAGGAA----------------------------------ATGGC---------------------TCTGCGTCCAGCTGATTGATTATGGGACAG-----------------A-------GGCATTCGCTTTG---------C-----C-----------------TGACA-ACCTTAATTAAAACAACTTTTTG-------GTTACCTT--------------------------TACAGCTGCT-C----------------------------------------------------------------------T---GCCACAACC---ACA---------ACAACAAC---------AACAACAGCA----ACAGTTTCTATAGTCGCGCAGAC---------------------ATCTGCAACATCAGCGTTTG------------------------------------------------------------GCCCA |
| droVir3 |
scaffold_13049:3218832-3219096 - |
|
GGT---------ATT--GGTGGCAGCCA---CATTGTAATAGAGCCAT-GCTGCATCCACGA---AAATGC------------------------TGCAGCAGGAA----------------------------------ATGG---------------------------------CTGATTGATTATGAGACAG----------------AA-------GGCATTCGCTTTTAAATAAACGCTTCTCT-----------------TGGCA-TTTTTAATTAAAGCAAGTTTC-G-------TTTGCCTT----------CTTCCATCTTACCCC---CAG-----------------------------------------------------------------CTGCAGTCTATGT---GCCACAACAACAACA---------ACAACAAC---------AACAGCAGCA----GCAGTTTTTATAGCCGCGCCGAC---------------------ATCTGCAACATCAGCGTTTG------------------------------------------------------------GCCCA |
| droGri2 |
scaffold_15110:4141839-4142108 - |
|
GGG---------GGATTGCTTTTAGCCCCGGCATTATAATAAAGACAT-G--------------------------------------------------CAGGAA----------------------------------ATAC--GCTGCTCTGCTCAGCTCTGCTCAGCTCCCAGCTGATTGATTATGAGTTT------------------T-------GGCATTCGCTTTT---------CTTTTCTTTTCATTTCTTTTCGATTTATA-ATTTTAATTAAACCAACTTTT-C-------TT--------------ATCCTCCA---------------------------------------------------------------------CTTTCCAACAGCTGCTGTCAATGT---G------CA---ACAACAACAACAACAATAAC---------A-------------GTCGCTTCTACAGTCGCGCTGAT---------------------ATTTGCAACATCAGCGTCTG------------------------------------------------------------GCCCA |
| droWil2 |
scf2_1100000004590:4385525-4385704 - |
|
GCA---------AGT----------------------------------G--------------------CAACAGCAGCAGA------------CACAGCAGGCA----------------------------------GTGGCTGCGGCCGCAGCCGCCG---------------------------------------------------------------------TCA---------C-----C-----------------CAGCA-GCTTCAGCAACAACAG-------------------------------------------------------------CAGGCCGTCGCCCAA---CAGCAGCAGCAACAG---------------CAG-----------------------------CA------------------ACAAC---------AACAGCAG----------------------CAGCAACAACAGCAGCAGCAGCAAGTGCAACAGCAACAGCAAGTAGTG------------------------------------------------------------GCCCA |
| dp5 |
XR_group3a:978635-978741 + |
|
TTA---------AAATTAC-AT------------------------------------------------------T---------------------------------------------------------------TTCCTCACTGCTCTCTCCT--------------------------------------------------------------------------------------------------------------------------------------------------CTCTCT--------------------------CTCAC--------------------------------TCGCTC---------------------------------------T---GCCAC-----------------------------------------CAGCA----GCA-TTTCTATAGCCGTGCGGAC---------------------ATTTGCAACATCAGCGTGTG------------------------------------------------------------GCCCA |
| droPer2 |
scaffold_27:146643-146713 + |
|
CCTGACCAACTCAAACGATTTGCTC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGTTA-----------------------------------------GCCGAC-A----------------------------------------------------------------------T---GACC----------------------------------------------------------------------AAAC---------------------AACAACAACAACAAAAACAG------------------------------------------------------------CAACA |
| droAna3 |
scaffold_13337:10678131-10678279 + |
|
TGA---------CAACA-TTTGCAGG------ATTCCAACAACATCATGGCCGC----ACCCAGCCAAAGC--------------------------------------------------------------------------CACAGCCACAGCCA--------------------------------------------------------------------------------------------------------------CCA--------CATCTATGA-------------------------------------------------------------CTTG-----------CCACAG-----------------------------------------------------------------------CAATTGCAGCAGC------------------A----ACAACATCTACAATTGCAACAAC---------------------AACAACAGCAACAGCATCAA------------------------------------------------------------CATCA |
| droBip1 |
scf7180000396426:303787-303872 + |
|
TGC---------TC---------------------------------A-G--------------------CAG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGC---AA---------------C--------------------------------------A---------ACAGCAAC---------AACAGCAGCAAAACGCAGTGCAAGCGGCGGCCCAAGC---------------------AGCCGCTGCAGCAGCAGCTG------------------------------------------------------------GTC-- |
| droKik1 |
scf7180000302476:715816-715943 + |
|
GGG---------CCA--------------------------------T-G--------------------CAA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGCAACAG---------------CA-----------------------------------------------GAAGCAGAAGCAGCAACA-------------------------------CCAGC---------------------AGCAGCAGCAGCAGCATCAGACTGCAAATGGAGTCGGGGGCAGCAACATTATCCTGCTGCGCGGCACTCGCAACGAAAATGGC-- |
| droFic1 |
scf7180000454073:2983433-2983554 - |
|
GGC---------ATT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGCCGTG---------G-----C-----------------TGTCGCACTTTCGGTAATGTGACACTTTG---------TGCCAT--------------------------GG-----------CATG-----------TGGCAG---CTGCAACAA---------------CAC-----------------------------CA------------------ACAAC---------ATCACCAGCA----G---------------CAGCTAC---------------------AACAACAACAACAACATT--------------------------------------------------------------GGCAA |
| droEle1 |
scf7180000491006:114939-115160 - |
|
GCA---------ACA--------------------------------A-G--------------------CAGCAGCAGCAGCAGCAACATCAGCAGCATC--------------------------------------------------------------------------AGCAGAAGAACGAGGAGTCCAAAGTGTTCATCGACATACGGAATTCAGCACCAG---------------------------------------------ACGTGATTATCATGACGTCCCA-------TTGACATTTAAGGATGGCCCGCCA---------------------------------------------------------------------ATCAGCAACAACAACGGTA-GTGAACAG------GA------------------GCAAC------------------A----ACAATAGCAACAACAGCAGCATC---------------------AACAACAACAGCAGCAGCAG------------------------------------------------------------GAGCA |
| droRho1 |
scf7180000770177:24415-24495 - |
|
ACA---------AC---------------------------------A-A--------------------CAACAAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AA---------------C--------------------------------------A---------ACAACAAC---------A-------------ACAACAACAACAGCTGCGCAACCAACAAC---------------AACATCAACAACAACAGCGG------------------------------------------------------------GAGCA |
| droBia1 |
scf7180000300910:514843-514894 - |
|
AGC---------A---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAGTTTCTACAGCCGCGCGGAC---------------------ATCTGCAACATCAGCGTGTG------------------------------------------------------------GCCAG |
| droTak1 |
scf7180000414529:16033-16122 + |
|
TAC---------AAACGCTTTGCTG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGCTTCCA--------------TCAG---CATCAGAAACAG---------------CAG-----------------------------CA------------------TCAAC---------A----------------------TCACCATCCCAAAC---------------------AGCTCCATCAGCAGCACAAG------------------------------------------------------------CCCCC |
| droEug1 |
scf7180000407600:19519-19597 + |
|
CAA---------TTTCT-TTGA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAAC---GT-----------------------------------------------CA---ACC---------GTAACAACAACAACAACC-------------GCAACTGTAACGACAGCGCGATT---------------------AACGACGTCATCAGC---------------------------------------------------------------------- |
| dm3 |
chr3R:20277838-20277908 - |
|
ACG---------ACGGCGC----------------------------A-G------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGCAACAT---------------CAG-----------------------------CA------------------GCAAC---------ATCAGCAG----------------------CAACAGC---------------------AACTGCAACATCAGCAACTG------------------------------------------------------------CAGCA |
| droSim2 |
3r:19809086-19809158 - |
|
CGC---------A-----------------------------------------------------------G-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGCAG--------------------------------------------------------------CAACAGCAGCAGCAGCAGCAACA----------------------TCAGCAGCAACAGC---------------------AATTGCAACATCAGCAACTG------------------------------------------------------------CAGCA |
| droSec2 |
scaffold_0:20614376-20614442 - |
|
CGC---------AGC--------------------------------A-G------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGCAGCAG---------------CAG-----------------------------CA------------------GCAAC---------ATCAGCAG----------------------CAACAGC---------------------AATTGCAACATCAGCAACTG------------------------------------------------------------CAGCA |
| droYak3 |
3L:10573659-10573795 + |
|
CAA---------CAGCA-GCAG------------------------------------------------------CAGCAGCAG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGCTGTTGTCCAG---CAGCAACAGCAGCAGGTTGCTGCCGCCGCCCAG-----------------------------CA------------------GCAAC---------ATCAGCAG----------------------CAGCAACAACAACAACAGCAGGCCGTGCAGCAGCAGCAGCAACAGGCG------------------------------------------------------------GTCCA |
| droEre2 |
scaffold_4929:14588571-14588761 + |
|
CAA---------GC---------------------------------A-A--------------------CAG---CAGCAGCAGCAACAACAACAGCAGCAGCAAAAGAATTTCGGGGGCATACGCTTTAATTTGAATTTAAACCAAAAGCGTTTGGC------------------------------------------------------------------------------------------------------------------------------C---------GGTGGTCCCA--------------------------ATCCCCTGCAGCAGCAGCA--------------GCAA---C---------AA-----------------------------------------------CA---ACA---------ACAACAGC---------AGCAACAGCA----ACAACTTCAACAACTTCAACAAC---------------------AACAACATCATCAGC---------------------------------------------------------------------- |