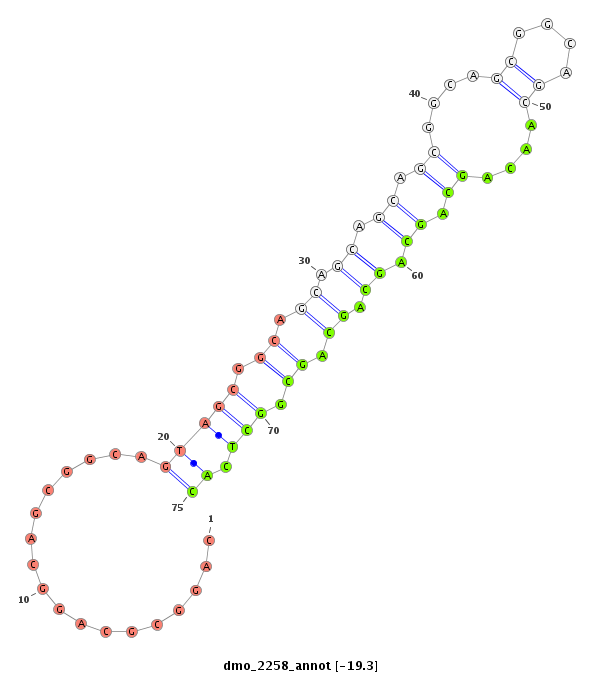
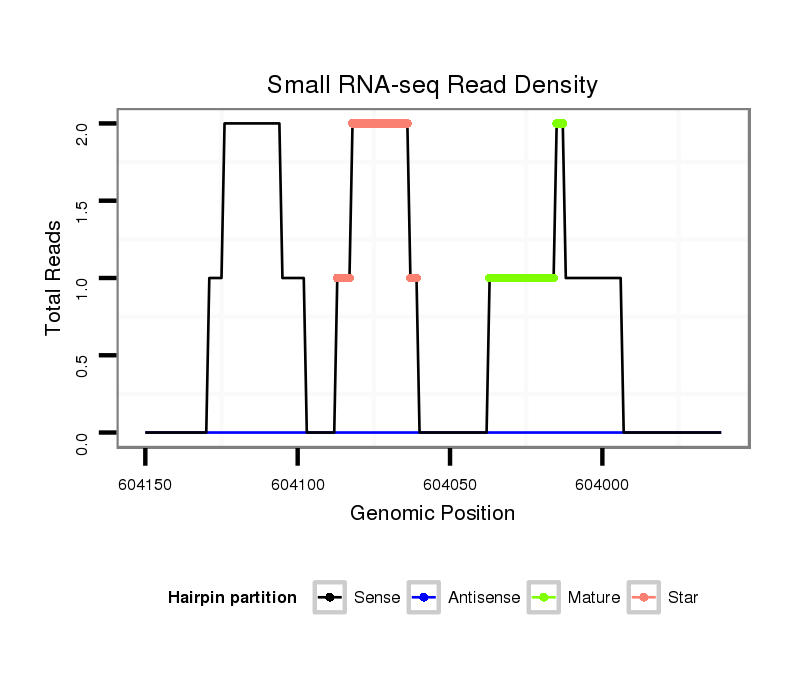
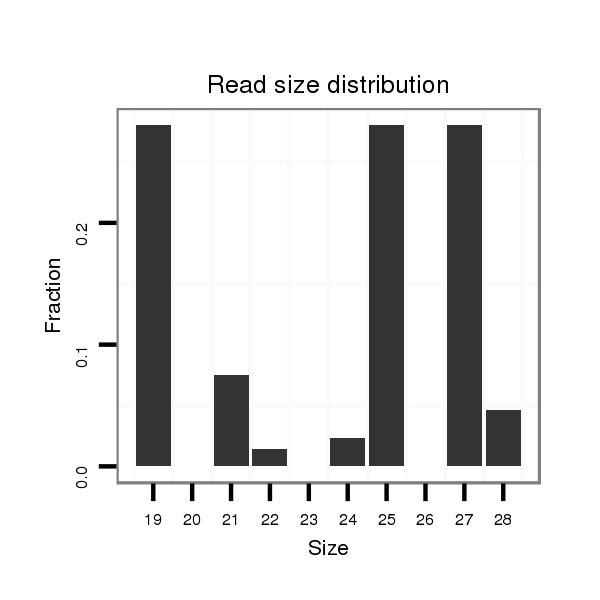
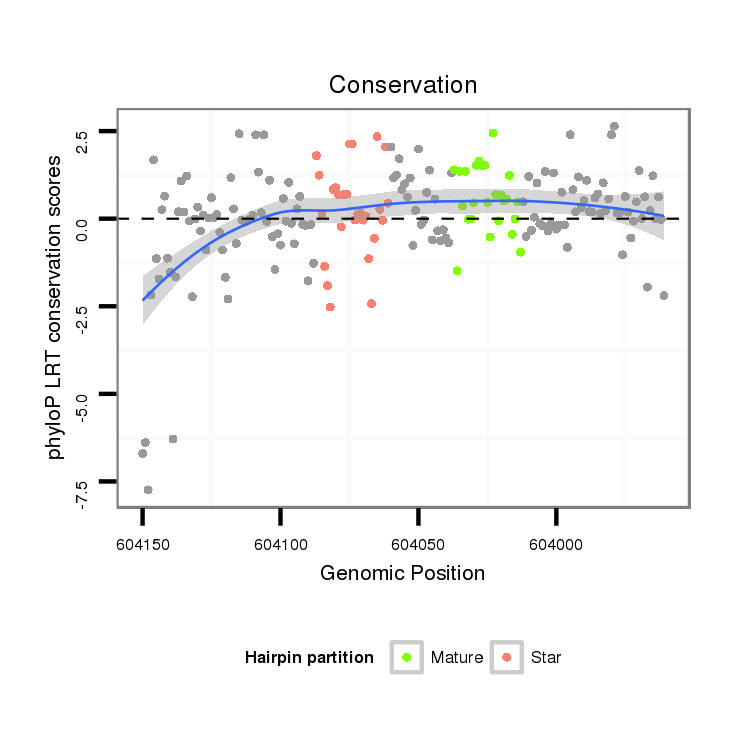
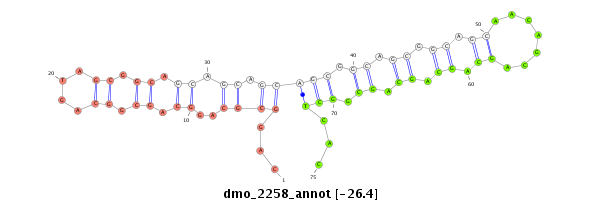
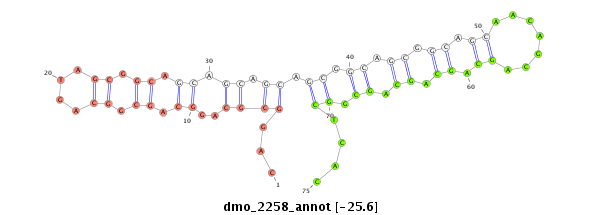
| TCG---------TATGGC---------GG---G-------------------------------------TACAGC-AGTGGCAGCGGG--------------------------------TGCAACGGCGGCAGCG---------------------------GTG-----------------------GTGGCAGCG--GGTACA---GCAG----T-G---GCATCGGGTGCAGCGGCGG-------------------------------------------TGGGTACAGCAGTGGCAG------C---GG------GT------------GCAGCAGTGGCAGCGGC---------------GGCAGGTACCGTAATGG---------------------------AT----GCGACA-------------------GCAGCGGCGG------CGGGTACAGCGGTGGC-------------AGCG---GCGGTG-- | Size | Hit Count | Total Norm | Total | M020
Head | M045
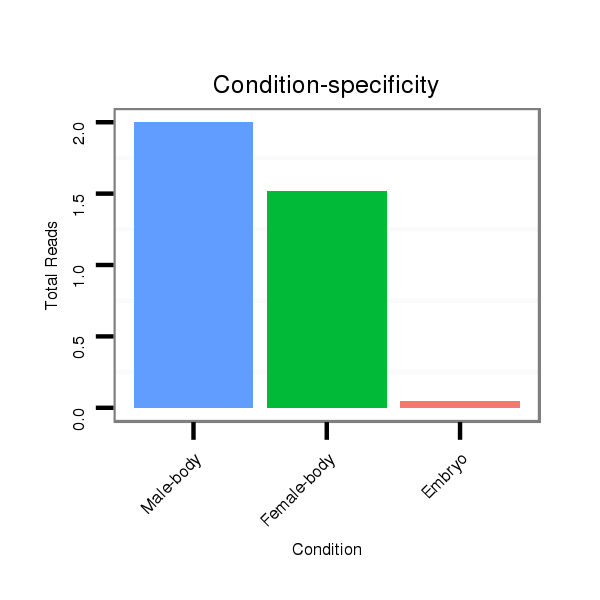
Female-body | V117
Male-body | V118
Embryo | V119
Head |
| .........................................................................................................................TGCAACGGCGGCAGCG---------------------------GTG-----------------------G........................................................................................................................................................................................................................................................................................................................ | 20 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 | 0 |
| ..............TGGC---------GG---G-------------------------------------TACAGC-AGTGGCAGCGG............................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................AT----GCGACA-------------------GCAGCGGCGG------CGG......................................... | 21 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................G-------------------------------------------TGGGTACAGCAGTGGCAG------C---GG------..................................................................................................................................................................................... | 22 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ................................................................................................................................GCGGCAGCG---------------------------GTG-----------------------GTGGCAGCG--G............................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................ATGG---------------------------AT----GCGACA-------------------GCAGCGGCG................................................... | 21 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................................................TGG---------------------------AT----GCGACA-------------------GCAGCGGCGG------C........................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................GGG--------------------------------TGCAACGGCGGCAGCG---------------------------G.................................................................................................................................................................................................................................................................................................................................................. | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................TG-----------------------GTGGCAGCG--GGTACA---GCAG----T-T---....................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................GCAGTGGCAGCGGC---------------GGCAGGA................................................................................................................................ | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................GT------------GCAGCAGTGGCAGCGGC---------------GG..................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................CAGCGGG--------------------------------TGCAACGGCGGC.................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................TGCAACGGCGGCAGCG---------------------------GTG-----------------------GTGG..................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................G-------------------------------------TACAGC-AGTGGCAGCGGG--------------------------------T............................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................AGCGGG--------------------------------TGCAACGGCGGCAGCG---------------------------G.................................................................................................................................................................................................................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................AGCAGTGGCAGCGGC---------------GGCAGGT................................................................................................................................ | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................TG-----------------------GTGGCAGCG--GGTACA---GCAG----T-G---G...................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................ACAGCAGTGGCAG------C---GG------GT------------GCA.................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................CAGCG---------------------------GTG-----------------------GTGGCAGCG--GGTACA---..................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................AGCGGG--------------------------------TGCAACGGCGGCAGT............................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................TACAGC-AGTGGCAGCGGG--------------------------------TGCAACGT...................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................GCAGCGGG--------------------------------TGCAACGGCGGCC................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................GCAACGGCGGCAGCG---------------------------GTG-----------------------GTG...................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................................................GCGGCGG------CGGGTACAGCGGTGGC-------------AG............. | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................CA-------------------GCAGCGGCGG------CGGGTACAG................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................C---------GG---G-------------------------------------TACAGC-AGTGGCAGC................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................................................G---------------------------AT----GCGACA-------------------GCAGCGGCGG------CG.......................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................T----GCGACA-------------------GCAGCGGCGG------CGGGT....................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................AGCGGG--------------------------------TGCAACGGCGGCAGC............................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |