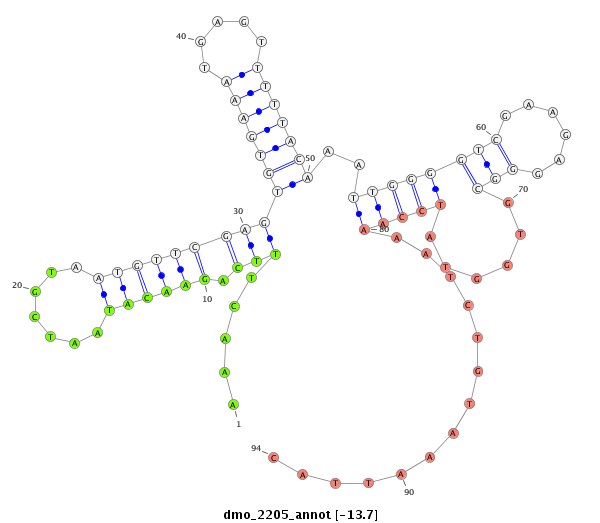
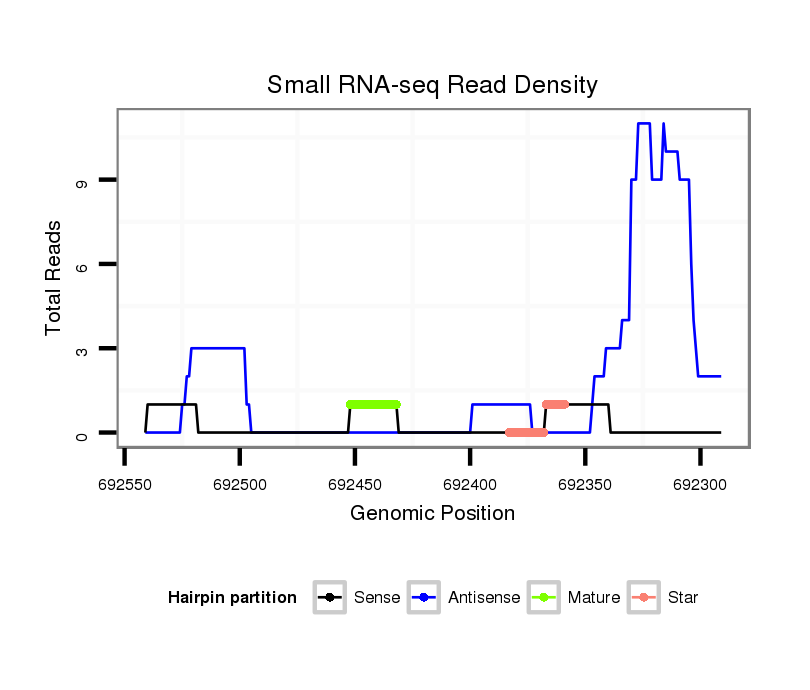
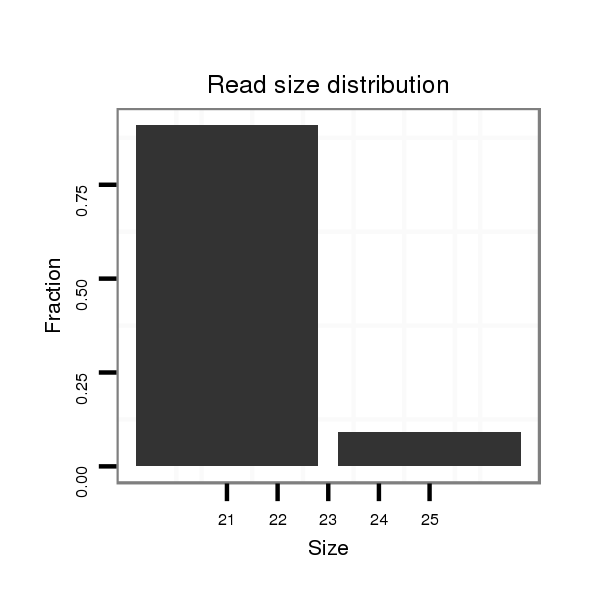
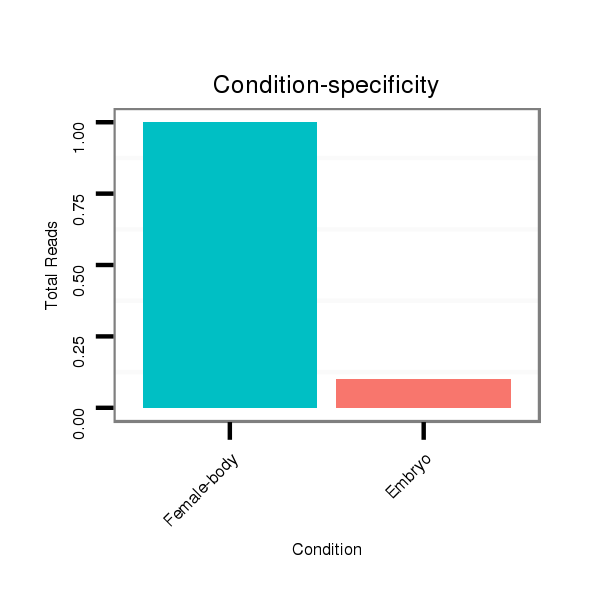
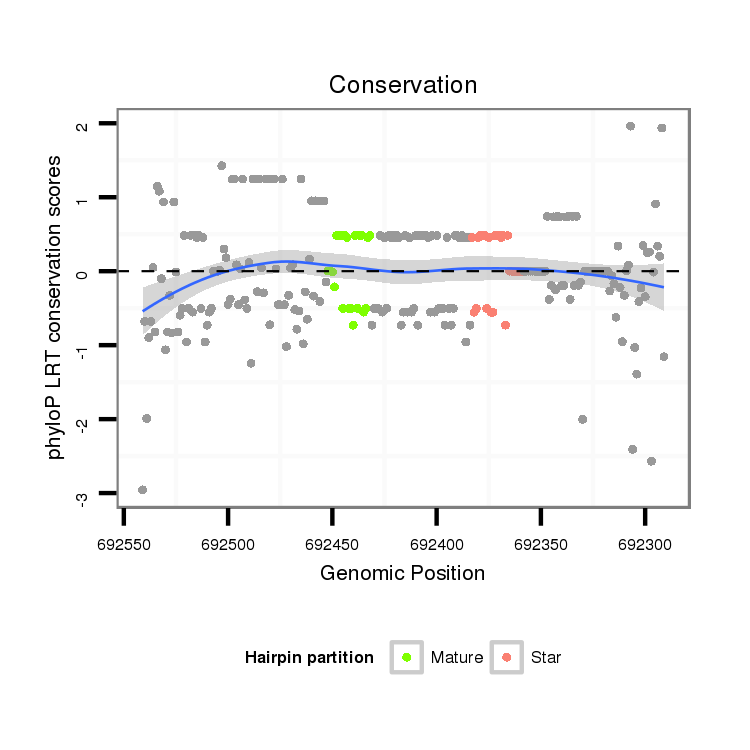
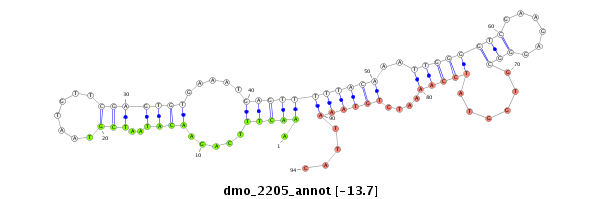
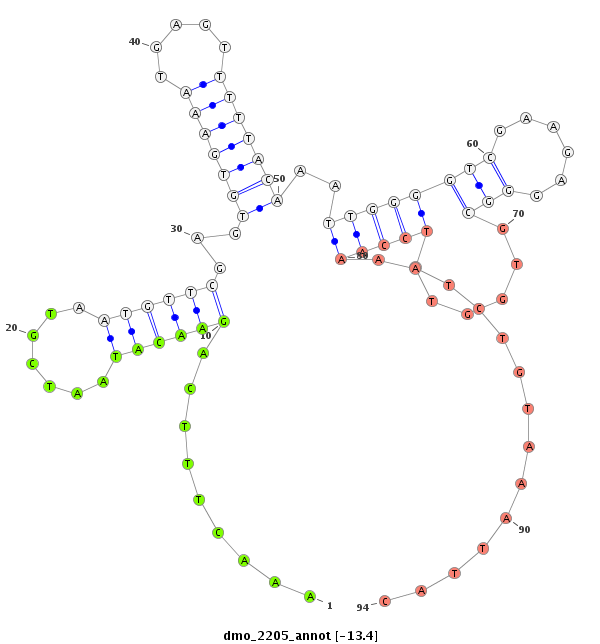
| GG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TG------------CGAACGACGA--------------------TCAGATTGGCGGATGGG- | Size | Hit Count | Total Norm | Total | M022
Male-body | M040
Female-body | M059
Embryo | M062
Head | V043
Embryo | V051
Head | V112
Male-body | SRR902010
Ovary | SRR902011
Testis | SRR902012
CNS imaginal disc |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGA..... | 23 | 16 | 27.00 | 432 | 1 | 302 | 3 | 0 | 2 | 1 | 0 | 102 | 21 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGG...... | 22 | 16 | 26.63 | 426 | 0 | 388 | 4 | 0 | 3 | 0 | 1 | 24 | 6 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGAT.... | 24 | 16 | 16.56 | 265 | 0 | 213 | 3 | 0 | 1 | 0 | 1 | 46 | 1 | 0 |
| ...................................................................................................................................................................................................................................................................TG------------CGAACGACGA--------------------TCAGATTGGCGGA..... | 25 | 1 | 8.00 | 8 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCG....... | 21 | 17 | 7.94 | 135 | 0 | 107 | 3 | 0 | 6 | 0 | 1 | 14 | 4 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGC........ | 20 | 17 | 7.24 | 123 | 0 | 69 | 4 | 0 | 8 | 0 | 1 | 28 | 13 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGATGG.. | 26 | 16 | 5.44 | 87 | 0 | 82 | 1 | 0 | 0 | 0 | 0 | 4 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGATG... | 25 | 16 | 4.63 | 74 | 0 | 65 | 0 | 0 | 0 | 0 | 0 | 8 | 1 | 0 |
| ....................................................................................................................................................................................................................................................................G------------CGAACGACGA--------------------TCAGATTGGCGG...... | 23 | 1 | 4.00 | 4 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................TG------------CGAACGACGA--------------------TCAGATTGGCG....... | 23 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................GAACGACGA--------------------TCAGATTGGCGG...... | 21 | 16 | 1.75 | 28 | 0 | 26 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................GAACGACGA--------------------TCAGATTGGCGGA..... | 22 | 16 | 1.31 | 21 | 0 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGG......... | 19 | 17 | 1.12 | 19 | 0 | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................TG------------CGAACGACGA--------------------TCAGATTGGC........ | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................................................G------------CGAACGACGA--------------------TCAGATTGGCGGA..... | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................G------------CGAACGACGA--------------------TCAGATTGGCG....... | 22 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TG------------CGAACGACGA--------------------TCAGATTGG......... | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................TG------------CGAACGACGA--------------------TCAGATTGG......... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................G------------CGAACGACGA--------------------TCAGATTGGC........ | 21 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................TG------------CGAACGACGA--------------------TCAGATTGGCGGAT.... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................TG------------CGAACGACGA--------------------TCAGATTGGCGG...... | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................TG------------CGAACGACGA--------------------TCAGATTGGCGGATG... | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................GAACGACGA--------------------TCAGATTGGCG....... | 20 | 17 | 0.76 | 13 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................GAACGACGA--------------------TCAGATTGGCGGATGG.. | 25 | 16 | 0.56 | 9 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGATGGG- | 27 | 16 | 0.56 | 9 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................GAACGACGA--------------------TCAGATTGGCGGAT.... | 23 | 16 | 0.56 | 9 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................GAACGACGA--------------------TCAGATTGGCGGATG... | 24 | 16 | 0.44 | 7 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGAC.... | 24 | 16 | 0.44 | 7 | 0 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGATA... | 25 | 16 | 0.38 | 6 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................AACGACGA--------------------TCAGATTGGCGGATGG.. | 24 | 16 | 0.31 | 5 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................GACGA--------------------TCAGATTGGCGGATGG.. | 21 | 16 | 0.31 | 5 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGAA.... | 24 | 16 | 0.25 | 4 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGT...... | 22 | 17 | 0.24 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGG..... | 23 | 16 | 0.19 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................GAACGACGA--------------------TCAGATTGG......... | 18 | 17 | 0.18 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................ACGA--------------------TCAGATTGGCGGATGG.. | 20 | 16 | 0.13 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGAG.... | 24 | 16 | 0.13 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................GAACGACGA--------------------TCAGATTGGC........ | 19 | 17 | 0.12 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGAAAA.. | 26 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGATGT.. | 26 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGAAA... | 25 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGATAC.. | 26 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................GAACGACGA--------------------TCAGATTGGCGGATA... | 24 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGACAA.. | 26 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................GAACGACGA--------------------TCAGATTGGCGGAA.... | 23 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................ACGACGA--------------------TCAGATTGGCGGATGT.. | 23 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGATGGT- | 27 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGATGGA- | 27 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................GAACGACGA--------------------TCAGATTGGCGGAAA... | 24 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................AACGACGA--------------------TCAGATTGGCGGA..... | 21 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................ACGACGA--------------------TCAGATTGGCGGAT.... | 21 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGATC... | 25 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................GAACGACGA--------------------TCAGATTGGCGGT..... | 22 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGATAA.. | 26 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGT..... | 23 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................................................................GACGA--------------------TCAGATTGGCGGA..... | 18 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................AACGACGA--------------------TCAGATTGGCGGATGA.. | 24 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCGGATGA.. | 26 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGCA....... | 21 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTG.......... | 18 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................CGAACGACGA--------------------TCAGATTGGTAT...... | 22 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |