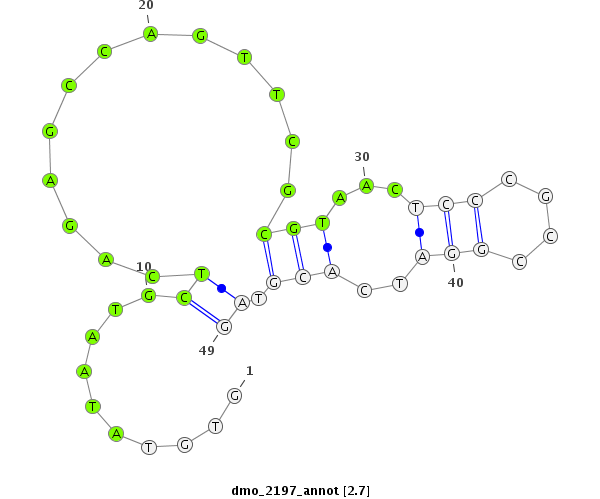
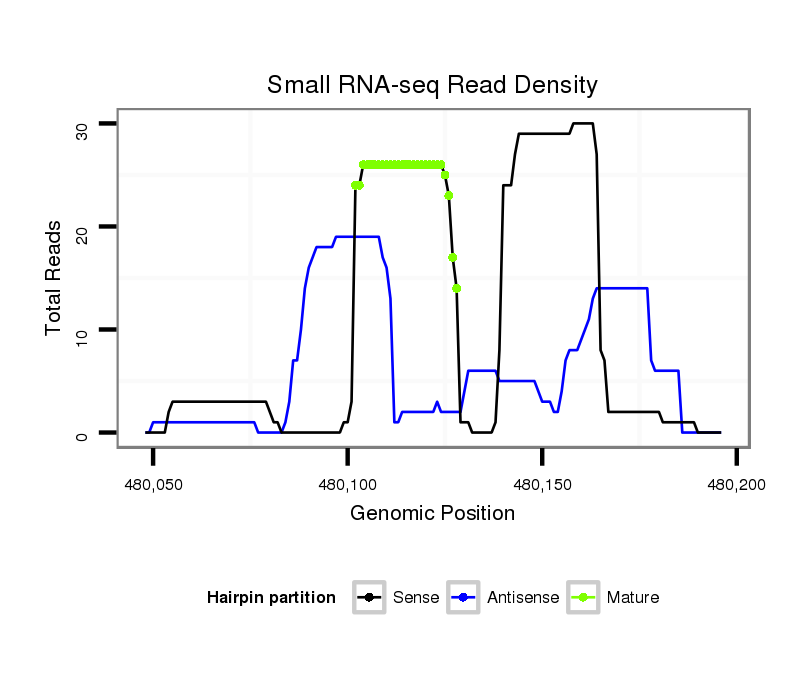
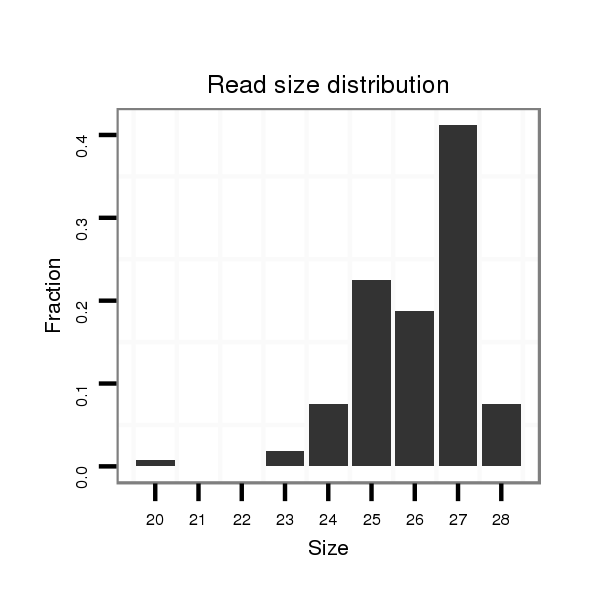

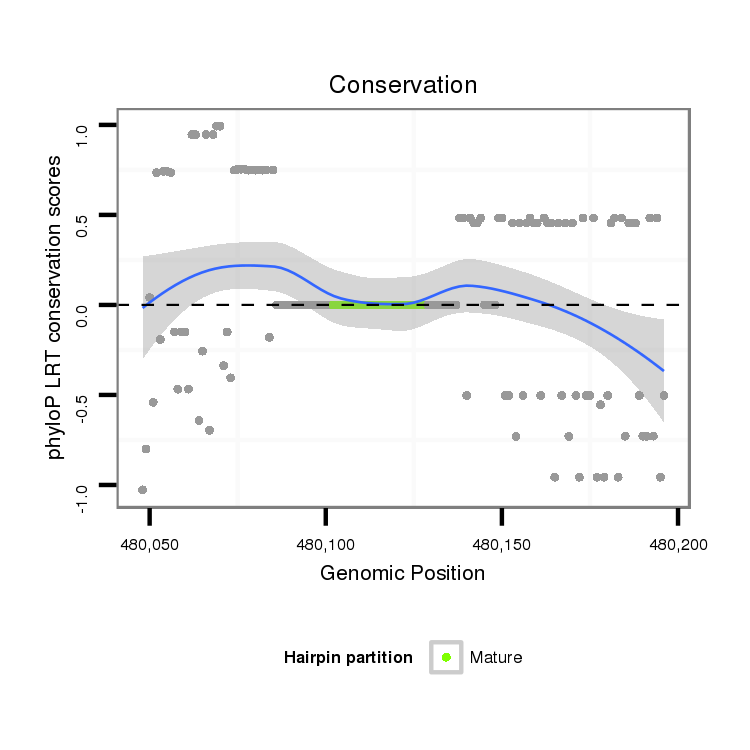
ID:dmo_2197 |
Coordinate:scaffold_6541:480098-480146 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -5.0 | -4.9 | -4.9 | -4.7 |
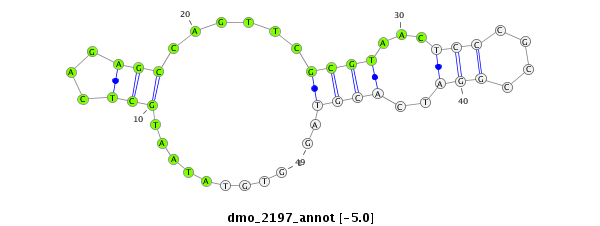 |
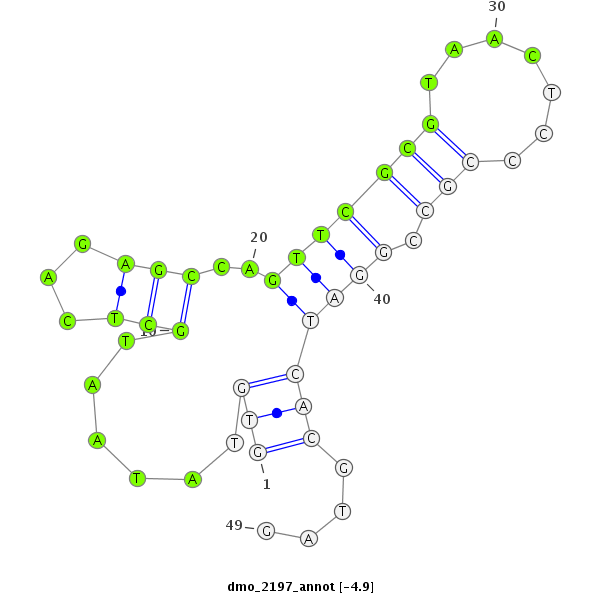 |
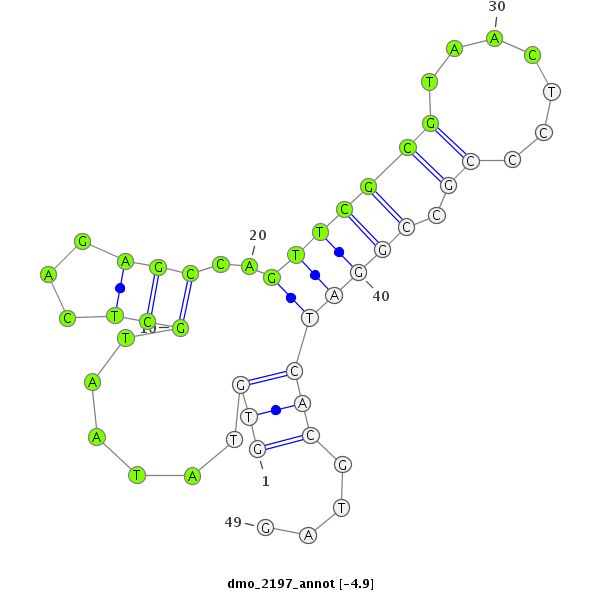 |
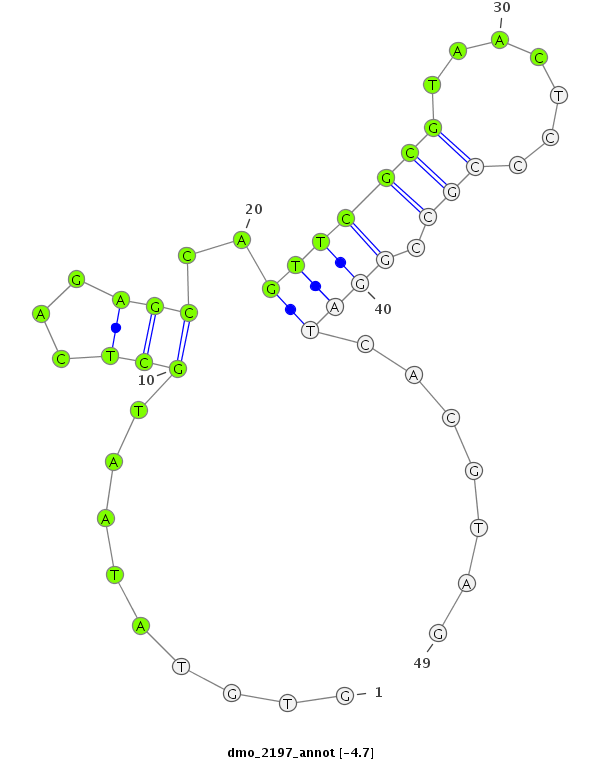 |
intron [Dmoj\GI14200-in]; CDS [Dmoj\GI14200-cds]; exon [dmoj_GLEANR_14475:2]; CDS [Dmoj\GI14200-cds]; exon [dmoj_GLEANR_14475:3]
No Repeatable elements found
| ##################################################-------------------------------------------------################################################## AACCCGTTCGCGCCCGCAGCCAAACGGATTGCGGCCACGATGCGGCCCTGGTGTATAATGCTCAGAGCCAGTTCGCGTAACTCCCGCCGGATCACGTAGAGATATCTGGGTGCCAGCGGTGTCTCACGTCAGACTCTGGCGTATAATGC **************************************************..........((.............(((...(((....)))..))).))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V110 male body |
V041 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................CACGTAGAGATATCTGGGTGCCAGC................................ | 25 | 0 | 1 | 14.00 | 14 | 9 | 5 | 0 | 0 |
| ......................................................ATAATGCTCAGAGCCAGTTCGCGTAAC.................................................................... | 27 | 0 | 1 | 11.00 | 11 | 1 | 10 | 0 | 0 |
| ......................................................ATAATGCTCAGAGCCAGTTCGCGTA...................................................................... | 25 | 0 | 1 | 5.00 | 5 | 2 | 0 | 2 | 1 |
| ...........................................................................................TCACGTAGAGATATCTGGGTGCCAGC................................ | 26 | 0 | 1 | 4.00 | 4 | 3 | 1 | 0 | 0 |
| ......................................................ATAATGCTCAGAGCCAGTTCGCGTAA..................................................................... | 26 | 0 | 1 | 3.00 | 3 | 2 | 0 | 1 | 0 |
| ...............................................................................................GTAGAGATATCTGGGTGCCAGCGG.............................. | 24 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 |
| ......................................................ATAATGCTCAGAGCCAGTTCGCGT....................................................................... | 24 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 |
| ..........................................................................................AACACGTAGAGATATCTGGGTGCCAG................................. | 26 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ...........................................................................................TCACGTAGAGATATCTGGGTGCCAG................................. | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ......TTCGCGCCCGCAGCCAAACGGATTGCG.................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................CACGTAGAGATATCTGGGTGCCAG................................. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........................................................AATGCTCAGAGCCAGTTCGCGTAACTCC................................................................. | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ...........................................................................................TCACGTAGAGATATCTGGGTGCCAGCGG.............................. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................................ATAATGCTCAGACCCAGTTCGCGTAA..................................................................... | 26 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ......TTCGCGCCCGCAGCCAAACGGATTGCGGC.................................................................................................................. | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......TCGCGCCCGCAGCCAAACGGATTGC..................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .....................................................TATAATGCTCAGAGCCAGTTCGCGTAAC.................................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................CACGTAGAGATATCTGGGTGCCAGCG............................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ..............................................................................................................TTCCAGCAGTGTCTCACGTCAGACTC............. | 26 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .....................................................TATAATGCTCAGAGCCAGTTCGCGTA...................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................................TGCCAGCGGTGTCTCACGTCAGA................ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...................................................TGTATAATGCTCAGAGCCAGTTCGCG........................................................................ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................ATCACGTAGAGATATCTGGGTGCCAGC................................ | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ..........................................................................................TTCACGTAGAGATATCTGGGTGCCAG................................. | 26 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................TAGAGATATCTGGGTGCCAGCGG.............................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................TCACGTACAGATATCTGGGTCCCAGC................................ | 26 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................................TCACGTAGAGATATCTGGGGGCCAGC................................ | 26 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........................................................AATGCTCAGAGCCAGTTCGCGTAAC.................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ........................................................................................................................GTCTCACGTCAGACTCTGGCGT....... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................................ATAATCCTCAGAGCCAGTTCGCGTAAC.................................................................... | 27 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ................................................................................................TAGAGATATCTGGGTGCCAGCGGT............................. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................CACGTAGAGATATCTGGGTGCC................................... | 22 | 0 | 3 | 0.67 | 2 | 0 | 2 | 0 | 0 |
| ........................................................................................................................GTCTCACGTCAGACTCTCTCC........ | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ......................................................ATAATGCTCAGAGCCAGTTCGCG........................................................................ | 23 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ......................................................TGAATGCTCAGAGCCAGTTCGCATAAC.................................................................... | 27 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ............................................................................................CACGTAGAGATATCTGGGTG..................................... | 20 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................TCACGTAGAGATATCTGGGT...................................... | 20 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................ACGTAGAGATATCTGGGTGCC................................... | 21 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ......................................................ATAATGCTCAGAGCCAGTTC........................................................................... | 20 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................GTCAGACTCTGGCGTATTTG.. | 20 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
|
TTGGGCAAGCGCGGGCGTCGGTTTGCCTAACGCCGGTGCTACGCCGGGACCACATATTACGAGTCTCGGTCAAGCGCATTGAGGGCGGCCTAGTGCATCTCTATAGACCCACGGTCGCCACAGAGTGCAGTCTGAGACCGCATATTACG
**************************************************..........((.............(((...(((....)))..))).))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V049 head |
V041 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................CCACGGTCGCCACAGAGTGCAG................... | 22 | 0 | 1 | 3.00 | 3 | 1 | 2 | 0 | 0 |
| .........................................CGCCGGGACCACATATTACGAGT..................................................................................... | 23 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 |
| ..................................................................................GGGCGGCCTAGTGCATCTCTATA............................................ | 23 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| .....................................GCTACGCCGGGACCACATATTACGAGT..................................................................................... | 27 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ......................................CTACGCCGGGACCACATATTACGAG...................................................................................... | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ..........................................GCCGGGACCACATATTACGAGT..................................................................................... | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ........................................ACGCCGGGACCACATATTACGAGT..................................................................................... | 24 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ...................................................................................................................CGCCACAGAGTGCAGTCTGAGAC........... | 23 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ....................................TGCTACGCCGGGACCACATATTACG........................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................CTACGCCGGGACCACATATTACGAGT..................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................GGCGGCCTAGTGCATCTCTATAG........................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................CCCACGGTCGCCACAGAGTGCAGT.................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ............................................CGGGACCACATATTACGAGT..................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................CGGTCAAGCGCATTGAGGGCGGCCT.......................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................GGTCGCCACAGAGTGCAGTCTGAGAC........... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .................................................................CCGGTCAAGCGCATTGAGGGC............................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .........................................CGCCGGGACCACATATTACGA....................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................................................GTCGCCACAGAGTGCAGTCTGAGAC........... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................CCCACGGTCGCCACAGAGTGCAG................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...........................................CCGGGACCACATATTACGAGT..................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................CACGGTCGCCACAGAGTGCAG................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................................TCGCCACAGAGTGCAGTCTGAGAC........... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .TGGGCAAGCCCGGGCGGGGGTT.............................................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ...........................................................................GCATTGAGGGCGGCCTAGTGCATCTC................................................ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................GCCACAGAGTGCAGTCTGAGAC........... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................GGCGGCCTAGTGCATCTCT............................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................CTACGCCGGGACCACATATTACG........................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................ACCCACGGTCGCCACAGAGTGCAG................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .................................................................................TGGGCGGCCTAGTGCATCTC................................................ | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| .................................................CCACATATTACGAGTCTCGGTCAAGCG......................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....................................TATACGCCGGGACCACATATTACGAGT..................................................................................... | 27 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..GGGCAAGCGCGGGCGTCGGTTTGCCTA........................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........................................ACGCCGGGACCACATATTACGAG...................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................GACCCACGGTCGCCACAGAGTGCAG................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| TTGGGCAAGCACGGGCGCGGGTT.............................................................................................................................. | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ...............CGTCGGTTTGCCTAACGCCGGTGCTA............................................................................................................ | 26 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ...........CGGGCGTCGGTTTGGCTCAC...................................................................................................................... | 20 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................CGGGCACGCGCGTTGAGGGC............................................................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6541:480048-480196 + | dmo_2197 | AACCCGTTCGCGCCCGCAGCCAAACGGATTGCGGCCACGATGCGGCCCTGGTGTATAATGCTCAGAGCCAGTTCGCGTAACTCCCGCCGGATCACGTAGAGATATCTGGGTGCCAGCGGTGTCTCACGTCAGACTCTGGCGTATAATGC |
| droVir3 | scaffold_12855:6849571-6849628 + | CGC---------------------------------------------------------------------------------------ATTACGT----ATGCCGGAGTGCTAGCCGCGGCCGATATGTCGCTGTTGCGCCGACTCT | |
| droKik1 | scf7180000299918:62660-62685 + | AGTGCATTCAAATACGAAGCCAAATG--------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000779914:222796-222827 + | TTC------GCGCCCGTGGGCAATCTGATTGCGGCCGC--------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
Generated: 05/18/2015 at 02:05 AM