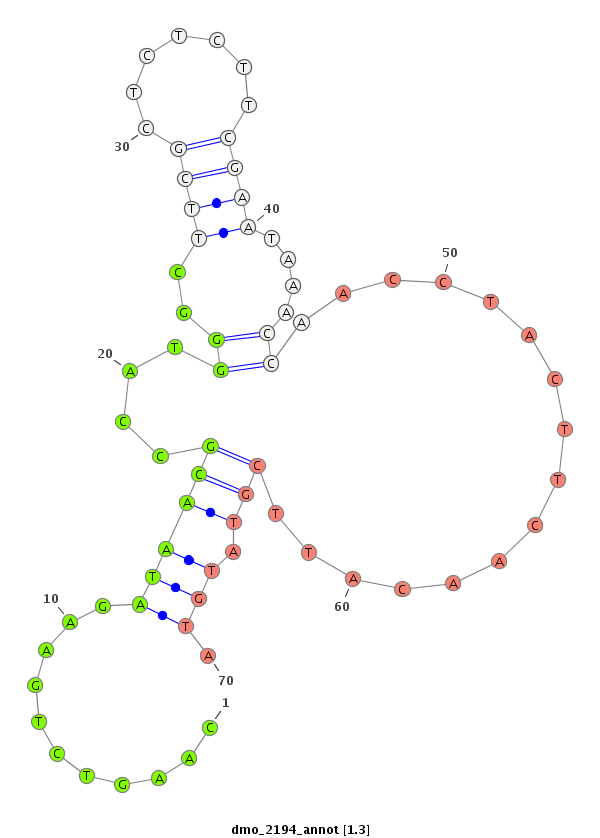
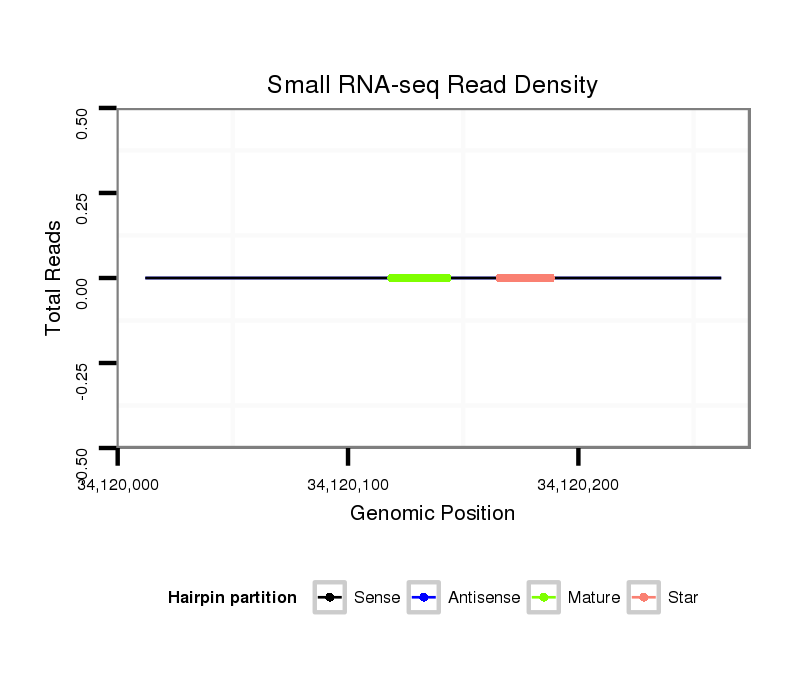
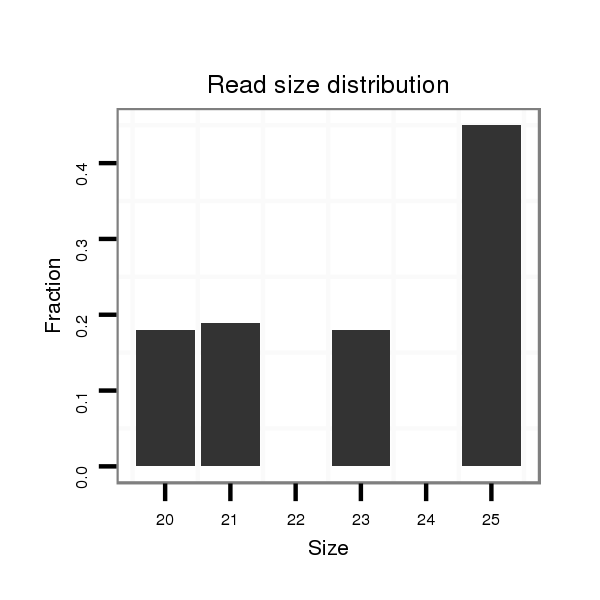
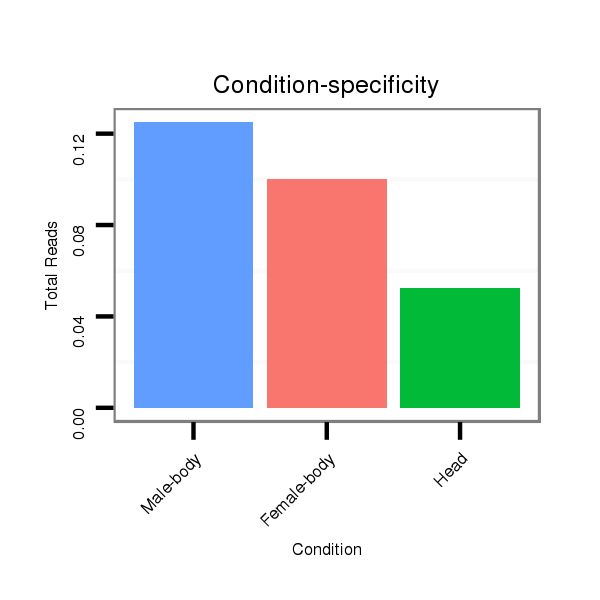
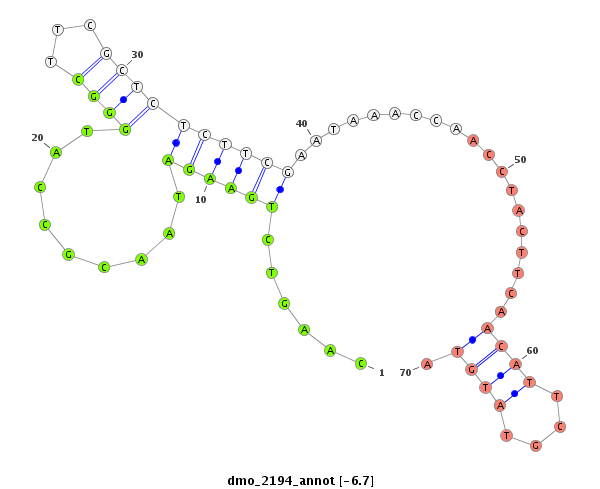
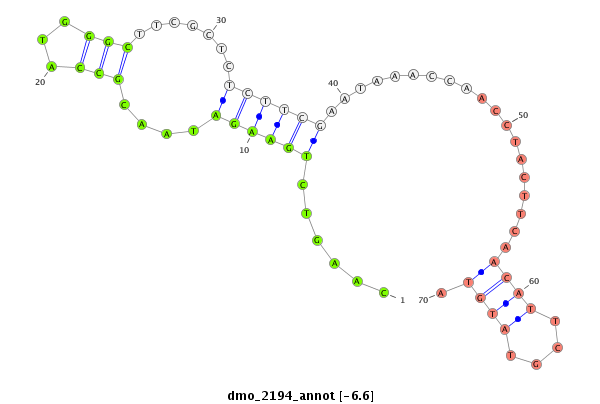
ID:dmo_2194 |
Coordinate:scaffold_6540:34120062-34120212 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
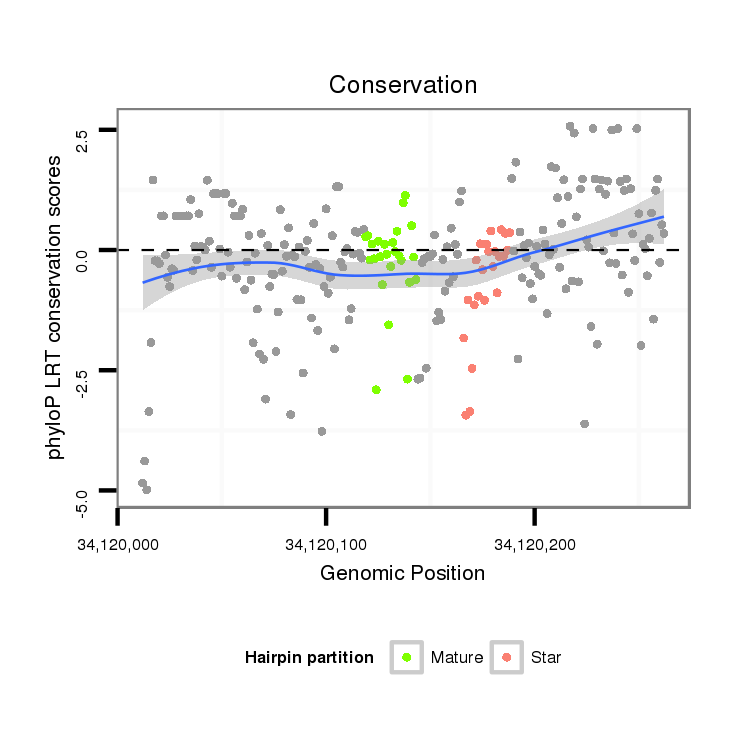
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -6.7 | -6.7 | -6.6 | -6.6 |
 |
 |
 |
 |
exon [dmoj_GLEANR_10753:6]; CDS [Dmoj\GI10860-cds]; intron [Dmoj\GI10860-in]
| Name | Class | Family | Strand |
| (CAA)n | Simple_repeat | Simple_repeat | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TGTTTAATATATTGTGTTAAAAACCAAAAAGACTAAAATAAGTGTGTTCTGCCCGCTGGTACCGGAGGTGCCGCTGCTGCGGCACCCGCAACAAAGAAGTAAGAGGCCAAGTCTGAAGATAACGCCATGGGCTTCGCTCTCTTCGAATAAACCAACCTACTTCAACATTCGTATGTACTAACCGTACGAAAACAACAACAGCAACAGCAGCAACAACAACAAGAACAACAGCAACCACGAAAGCAGCAACA ***********************************************************************************************************...........((((((....((..((((.......))))....))................))).))).************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V110 male body |
V056 head |
M060 embryo |
V049 head |
|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................CAAACCTACTTGAACATGCG................................................................................ | 20 | 3 | 20 | 1.50 | 30 | 6 | 17 | 6 | 0 | 1 |
| .......................................................................................................................................................CAAACCTACTTGAACATGC................................................................................. | 19 | 3 | 20 | 0.90 | 18 | 18 | 0 | 0 | 0 | 0 |
| .................................................TGCCCGCTGGTACCGGAGGTGCCGCTG............................................................................................................................................................................... | 27 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................AACCTACTTGAACATGCG................................................................................ | 18 | 2 | 20 | 0.15 | 3 | 2 | 0 | 1 | 0 | 0 |
| ...........................................................................................................CAAGTCTGAAGATAACGCCATGGGC....................................................................................................................... | 25 | 0 | 16 | 0.13 | 2 | 0 | 2 | 0 | 0 | 0 |
| ..............................GACTAAAATAAGTGTGTTCTGCCCGC................................................................................................................................................................................................... | 26 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................ACGAAAACAACAACAGCAACAGC........................................... | 23 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................ACGAAAACAACAACAGCAACAG............................................ | 22 | 0 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................AACATGCGCATCTACTAAC..................................................................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................CGCAACAAAGAAGTAAGAGGC................................................................................................................................................ | 21 | 0 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................AAAGACTAAAATAAGTGTGTTCTGCCC..................................................................................................................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................AGAGCAACAGCAGCAACAACAACAAGG........................... | 27 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................GGCTTCGCTCTCTTCGAATA...................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................AACAACAACAGCAACAGCAGCAAC..................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................CTACAATAAGTGTGTTCC......................................................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................AACAACAGCAACAGCAGCAACAA................................... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................AACAGCAACAGCAGCAACAACAACA.............................. | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................CAAACCTACTTGAACATGCGT............................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................AAAAGACTAAAATAAGTGTGTTC.......................................................................................................................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................ACCTACTTCAACATTCGTATGTA.......................................................................... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................CTACAATAAGTGTGTTCCA........................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
|
ACAAATTATATAACACAATTTTTGGTTTTTCTGATTTTATTCACACAAGACGGGCGACCATGGCCTCCACGGCGACGACGCCGTGGGCGTTGTTTCTTCATTCTCCGGTTCAGACTTCTATTGCGGTACCCGAAGCGAGAGAAGCTTATTTGGTTGGATGAAGTTGTAAGCATACATGATTGGCATGCTTTTGTTGTTGTCGTTGTCGTCGTTGTTGTTGTTCTTGTTGTCGTTGGTGCTTTCGTCGTTGT
**************************************************************************...........((((((....((..((((.......))))....))................))).))).*********************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V110 male body |
V056 head |
V049 head |
M060 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................ACCATGGCCTCCACCCCAA................................................................................................................................................................................ | 19 | 3 | 20 | 2.55 | 51 | 50 | 1 | 0 | 0 | 0 |
| .............................................................GGCCTCCACGGCGACCGCGCTGT....................................................................................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................ATGATTGGCATGCTTTTGTTGTT..................................................... | 23 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................TTGGCATGCTTTTGTTGTTGTCGT................................................ | 24 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................ATTGCGGTACCCGAAGCGAGAGAAGCT......................................................................................................... | 27 | 0 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................GTTTGGATGAACTTGTACG................................................................................. | 19 | 3 | 20 | 0.35 | 7 | 0 | 6 | 1 | 0 | 0 |
| ........................................................................................................................TTGCGGTACCCGAAGCGAGAGAAGCTT........................................................................................................ | 27 | 0 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 | 0 |
| .......................................................................................................................ATTGCGGTACCCGAAGCGAGAGAAGCTT........................................................................................................ | 28 | 0 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 | 0 |
| ...................................................................................TGGGCGTTGTTTCTTCATTCTCCGGTT............................................................................................................................................. | 27 | 0 | 19 | 0.21 | 4 | 4 | 0 | 0 | 0 | 0 |
| ...................................................................................TGGGCGTTGTTTCTTCATTCTCCGGT.............................................................................................................................................. | 26 | 0 | 19 | 0.16 | 3 | 3 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................GTTTGGATGAACTTGTACGC................................................................................ | 20 | 3 | 20 | 0.15 | 3 | 1 | 0 | 0 | 2 | 0 |
| ..........................................................................................................................GCGGTACCCGAAGCGAGAGAAGCTTATT..................................................................................................... | 28 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GCGGTACCCGAAGCGAGAGAAGCTTATTT.................................................................................................... | 29 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 |
| ...........................................................................CGACGCCGTGGGCGTTGTTTCTTCATTCT................................................................................................................................................... | 29 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GCGGTACCCGAAGCGAGAGAAGCT......................................................................................................... | 24 | 0 | 20 | 0.15 | 3 | 2 | 1 | 0 | 0 | 0 |
| .........ATAACACAATTTTTGGTTTTTCTGATTT...................................................................................................................................................................................................................... | 28 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GCGGTACCCGAAGCGAGAGAAGCTTAT...................................................................................................... | 27 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 |
| .....................................................GCGCCCATGGCCGGCACGGC.................................................................................................................................................................................. | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................ACGGACGACCTTGGCCTC........................................................................................................................................................................................ | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................GGGCGTTGTTTCTTCATTCTCCGGT.............................................................................................................................................. | 25 | 0 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................ACCCGAAGCGAGAGAAGCTTATTTGGTT................................................................................................ | 28 | 0 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 | 0 |
| ....................................................................................GGGCGTTGTTTCTTCATTCTCCGGTT............................................................................................................................................. | 26 | 0 | 19 | 0.11 | 2 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TGCGGTACCCGAAGCGAGAGAAGCTTAT...................................................................................................... | 28 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..............................................................................CGCCGTGGGCGTTGTTTCTTCATTCT................................................................................................................................................... | 26 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................CGGTACCCGAAGCGAGAGAAGCTTATTT.................................................................................................... | 28 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................TTGGATGAACTTGTACGC................................................................................ | 18 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................GTTCTTGTTGTCGTTGGTGCTTTCGT...... | 26 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..........................................................................ACGACGCCGTGGGCGTTGTTTCTTCAT...................................................................................................................................................... | 27 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................ACCCGAAGCGAGAGAAGCTTATTTGGT................................................................................................. | 27 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| .............................................................................................................................GTACCCGAAGCGAGAGAAGCTTATTT.................................................................................................... | 26 | 0 | 20 | 0.10 | 2 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................CGGTACCCGAAGCGAGAGAAGCTT........................................................................................................ | 24 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................TGATGGGCATACTTCTGTT........................................................ | 19 | 3 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................CTGTTTCTTCATTCTCCGGTTCAGACTTC..................................................................................................................................... | 29 | 1 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................CCTGGGCGTTGTTTCTTCATTCTCCGGTT............................................................................................................................................. | 29 | 1 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................CGAAGCGAGAGAAGCTTATTTGGTT................................................................................................ | 25 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................GTGGGCGTTGTTTCTTCATTCTCCGGTT............................................................................................................................................. | 28 | 0 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................TTGCGGTACCCGAAGCGAGAGAAGCTTAT...................................................................................................... | 29 | 0 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ACAAATTATATAACACAATTTTTGGTTTT.............................................................................................................................................................................................................................. | 29 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............CAATTTTGGGTTTTTCTGATT....................................................................................................................................................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................GTTGGTGCTTTCGTCGTTGT | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TTGTCGTTGGTGCTTTCGT...... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................GAAGCTTATTTGGTTGGATGAAGT....................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................ATTGCGGTACCCGAAGCGAGAGAAGC.......................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................GTTGTCGTTGGTGCTTTCGT...... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................AGAGAAGCTTATTTGGTTGGATGAAGTT...................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................TTCGGTACCCGAAGCGAGAGAAGCT......................................................................................................... | 25 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................ACCCGAAGCGAGAGAAGCTTATT..................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........ATAACACAATTTTTGGTTTTTCTGAT........................................................................................................................................................................................................................ | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................GACGCCGTGGGCGTTGTTTCTTCATT..................................................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................CTTGGTTGGATGAAGTTGTAAGCATA............................................................................. | 26 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................CGACGACGCCGTGGGCGTTGTTTCTT......................................................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................TACCCGAAGCGAGAGAAGCTTATTT.................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................TGGTTTTTCTGATTTTATTCACACA............................................................................................................................................................................................................ | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................CTTGTTGTCGTTGGTGCTTTC........ | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TGCGGTACCCGAAGCGAGAGAAGCT......................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................CTTGTTGTCGTTGGTGCTTTCGT...... | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................CGGTACCCGAAGCGAGAGAAGCTTATT..................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................TGTTGTTGTTCTTGTTGTCGTTGGT.............. | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................CGACGCCGTGGGCGTTGTTTCTTCATT..................................................................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................TGTTCTTGTTGTCGTTGGTGCTT.......... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................CACGGCGACGACGCCGTGGGCGTTGT.............................................................................................................................................................. | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................TTTGGTTGGATGAAGTTGTAAGCAT.............................................................................. | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................TGCGGTACCCGAAGCGAGAGAAGCTTATTT.................................................................................................... | 30 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................CGACGCCGTGGGCGTTGTTTCTTCAT...................................................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................GTTGTTCTTGTTGTCGTTGGTGCTT.......... | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................CCCGAAGCGAGAGAAGCTTATTTGGT................................................................................................. | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TGCGGTACCCGAAGCGAGAGAAGCTT........................................................................................................ | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GTTGTCGTCGTTGTTGTTGTTCTT.......................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................TTCTTGTTGTCGTTGGTGCTTTCG....... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................................................................................................................................TCTTGTTGTCGTTGGTGCTTTCGT...... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................ACGCCGTGGGCGTTGTTTCTTCATTCT................................................................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................GGTTTTTCTGATTTTATTCACACAAGA......................................................................................................................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................TGTTGTTGTTCTTGTTGTCGTT................. | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................TTGCGGTACCCGAAGCGAGAGAAGCT......................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................ACCCGAAACGAGAGAAGCTTATTTGGT................................................................................................. | 27 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................CGTCGTTGTTGTTGTTCTTGT........................ | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6540:34120012-34120262 + | dmo_2194 | TGTTTAATATATTGTGTTAAAAACCAAAAA-----G-----ACTAAAATAAGTGTGTTCTGCCCGCTGGTACC-----GGAGGTG----CCGCT--GCTGCGGCAC----------------CCGCAA----------------CAAA--GA-------------------------------------------------------------AGTAAGAGGCCAA---------------G-TCTGAAGATAACGCCATGGGCTTCGCT------------------------------------------------------------------------------------C-------------------------------------------------TCTT--------------CGAATAAACCAA-------------CCTACTTCAACATTCGTA---TGTACTAAC------------------------------------CGTACGA---AAACAA------CAA---------C--------------------------------------AG---CAACAGCA---------GCAACAACAACAAGAACAACAG------CAACCACG------A------------------AAGCAG---------CAA-CA |
| droVir3 | scaffold_12928:6658109-6658333 + | ACCAAA----------------------AA-----GAAGAAACCAAAGAAA-------CAGCGCGCCGTCACC-----ATGGATG-ATGCGGCC--GTTGCGGCAGCGG-------CAACAACTGGCG---------------------------------------------------------------------------G------------------------CAAT-------------TTGCAGCGCATGCAACAGAT--------------------------AACCA---------------------------AATCAGCATCGTCCAACATGATGTCACAGCG--------------------------------------------TTT-----------------------------------------------GCGC---A-------------------GCA--ACATGGA------------------------------TTACTACAACAA------T--------------------------------------------------AG---CA---------------GCAGCAGCAGCAACAACAACAGCAACAACAACAACT------A---------------CTA-TGCAG---------CAC-AT | |
| droGri2 | scaffold_15110:19327724-19327918 - | TGCTC------------------------------------------------------CTCCAGCTGGCACC-----GC------------------TTTGCCGC---CCGCCA-CGCCCATTACAG----------------T-----GA-------------------------------------------------------------CGCCAACGCCCGC--GCAGCA--------------TGATGCCAGCAATGGCTATACG----------------------------------------------------------------------------------------------------------------------------------------CC--------------GAGCTATAACAA----------------------------TCTAACGTCACCAGCATT------G------------------------AGCGCACAA---CAACAG------CAG---------C--------------------------------------AG---CAGCAGCA---------ACAACAACAGCAGCAGCAACAG------CAACAGCA------G------------------CAGCAA---------CAA-CA | |
| droWil2 | scf2_1100000004516:2951318-2951591 + | TGATAAGCTTATGTTTTTAAAAACAA---------------AGTAAAAAAAAGGTCTTCT-----TTG---------------TAATTGTTGTT--GTTGCTGATG----------------CTGCTGT-GTCCAGTTGTGACACAAA--GTGGGATAGAGAGCAAATGAGAGGGAGAGAAAAAA---------------------------------------------------------------------CAGCACAAGGTTTGCT----------------------------AAGGCGGCAA--C-------------------------------------------------GCAACGGCG------------------------------------CTT--------------GGGTTATAGCAA-------------TTTAC---A-------------------ATA--ACAGCAA-----------CTAC----------------------ACCAT----------------------------------------ACTATGCATATAATAAAAG---CAACAACA---------ACAACAACAACAACAACAACAA------CAACAACA------A------------------CAACAA---------CAA-CA | |
| dp5 | 3:13831454-13831705 + | GTGTAA-----CACG---------CCG----------AGAAACCAAAATAAATG------------------------TGTGGCAGTGGCAACA--GCGGTGACTG----------------CTGCTAGT---------------T--AT-----------------------------------------------------A--------------G---------CTTCTAGTTCCAAG-TGCAGCGACGACAACAGCAGCAACATCTGTAGAGACAACAAAAGAAGCCACC--------------A------------------------------CTAGTGTTAGAATA--------------------------------------------AAT-----------------------------------------------GGCC---T-------------------GCA--ACATGGCTTTGAATCCGGTATT---------A--ACAAAA---GAAGAG------GAA---------C--------------------------------------AC---CAGCAACA---------GCAACAGCAACAGCAACATCTG------CAACACCA------G------------------GAGTTT---------CAA-CA | |
| droPer2 | scaffold_13:1559169-1559361 - | GTGC-------------------------------------------------------TGCCCACAGTGATC-----AT----------CGAT--GATGCTGGTG----------CTTCAATTACAG----------------CCAA--CC-------------------------------------------------------------CGAAGGCCGCCAC-------------------------GGATAAGGCCAATGCCACC------------------------------------------------------------------------------------T-------------------------------------------------------------------CG-ACACCCAAA-------------ACGACGTCGACTCCTGGA---GGCGCCACCGCT------G--------C---------------------CAC---CGCCAA------TAG---------C--------------------------------------AG---CAGCAGCA---------GCAGCAACACCAACAGCCACAA------CATCAGCA------G------------------CAGCAG---------CAA-CA | |
| droAna3 | scaffold_13340:4279281-4279469 - | TGG------------------------------------------------------------CAGCGGCAGT-----GTCGCTGGCTGTGGCA--GTCACAACAT----------------TGT----------------------------------------------------------------------------------------CGCCGGCAGTCAT-----------------------------------------------------------------GACCAGCTCG----------------------------------------------------------------------------------------------------------CCAATGTCGCTGTCATGCATCCAACAT--------ACGGC--AGTTGCCAC------------------------AACAA---------------------------------------CAATAGCAACAG---------C--------------------------------------AA---CAGCAACA---------GCAATAACAACAACAACAGCAG------TAGCAGCA------G------------CAG---CCACAACAGCAATTGCAATCA | |
| droBip1 | scf7180000396543:349283-349493 + | GCAGGA----------------------GC------------------------------------------A-----GGAGGAG----CAGGA--GC----------------------------------------------------------------------------------AGGAGCGGCACAAGGTGCAACAA----------CGG----------------------------------GCGGCAGCAATAGCCACAGTTTAGAAGAGAGCAAAAGACTTGACC-----------------------------------------------------------A-------------------------------------------------TCAACAACAACAACAATATACAT----CAATATTACACACGGCCTTAATGTCGC---------------AACG---------------------------------------------------------------------------TTCTATG-------------------------CCACCTGCA---ACATCA------CCAGTTGCAACAACAACAACAG------CAACAACA------A------------------CAGCAA---------TA---A | |
| droKik1 | scf7180000302724:439655-439841 + | GCC------------------------------------------------------------AGGTGCAAAC-----GCAGGTGGCGGCAGCT-----GCAGCAG----------------CAGCCG---------------------------------------------------------------------------C------------------------ACAGTTGCAGCAGG-TTC------AACAGCAGCAGCATCATC------------------------------------------------------------------------------------A-------------------------------------------------------------------TGTACAAACCAC-------------ACAGATTCAGCACATTCA---AGTGCAATC------------------------------------TGCACCG---CCACCG------CAG---------C--------------------------------------AA---CAACAGCA---------GCAACAACAGCAGCAGCAGCAG------CAACAACA------G------------------CAGCAG---------CAG-CA | |
| droFic1 | scf7180000451454:2333-2549 - | AAAAA------------------CTAAAACAAAAAAAACAAAC----------------CATCTGCGAGAATC-----AT----------C--------GTGCCGC---TGATGATCGGCACTATCGG----------------CAAA--TAAG--AAAAAAGCT-ATAAGAGTGACATAGAAAA------------CATCAG----------CAGCAGCAAC---AATCATCAGCAAAAA---------------------------------------------------------------------------------------------------------------------------------------------------------------T-----------------------------------------------CTTC---A-------------------GCA--GCATCAG-----------CAGCAGCGGCAACAA-------------CA----------------------------------------------------------------ATCAGCA---------GCGGCAGCAACAACAACAACAA------CGGCAACA------A------------------CAACAG---------CAG-CG | |
| droEle1 | scf7180000490751:1099620-1099786 - | CCG----------------------------------------------------------------------------TCGGTG-TCGCAGCA--ACAGCAGCAG----------------CAGCAGC-------------------AG-----------------------------------------------------C----------------------A-ACAGCAGCAACAGG-CGCAACAG------------TT-----------------------------------------------------------------------------------------------GCCGCCGGG------------------------------------CGT--------------GGGAATTGCCAG-------------TCCGC---T-------------------GCA--ACAGCAG-----------CAGC----------------------AGCAA------CAG---------C--------------------------------------AG---CAGCAACA---------GCAGCAACAGCAACAGCAACAG------CAGCAACA------G------------------CAGCAG---------CAA-CA | |
| droRho1 | scf7180000780062:215807-215990 + | TCC------------------------------------------------------------ACCAACCACT-----ACCTC-TTGCGCGACC--ACCACAACAA----------------CCACAA-----------------------------------------------------------------------------------------------------CAACAACTAAAAA-GCCAACAACAACCACAACAACGACAAC---------------------A--------------------------------------------------------------A-------------------------------------------------------------------CTTGTAAACCAA-------------CAACAACCACAACA-------------------------A--------------C------GACAA--CAA-----------------C----------------TTGTAAA-------------------------CCAA---CAACAGCC---------TCAACAACAACAACAACCACAA------CAACAACA------A------------------CCACAA---------CAA-CT | |
| droBia1 | scf7180000301703:104024-104194 - | GGAGA----------------------------------------------------------------------------------TGCAGCA--GCTGCGCCGC---------------------------------------------------------------------------------------------------------------------------CAACAGCAGCAGG-GGCAGCAGT------------C-----------------------------------------------------------------------------------------------GCAT-----------------------------------------------------------------------------------TTCGG---G-------------------GCA--TCAGG-G------------------------GACGAGGAT---CACGAG------CAGAAACAACAGCAAC-AGGCACATAACCAA---CCACTACAAC-------CA---ACACAGCA---------CCACCAACTACAACCACAACAA------CAGCAACA------A------------------CAGCAA---------CAA-CA | |
| droTak1 | scf7180000415769:659481-659657 - | ATC---------------------------------------------------------------------------GCAGGTGGTGGCAGCT--GCCGCAGCGG----------------CCG------------------------------------------------------------------------------C------------------------CCAGCAGTTGCAAG-TTCAGCAGCAACAGCAACAGCATCACC------------------------------------------------------------------------------------A-------------------------------------------------------------------TGTACAAACCAC-------------CCAGATTCAGCACATTCA---AGTGCAATC------------------------------------TGCACCG---CCCCCG--------------------------------------------------------------CAACAACA---------GCAGCAGCAGCAACAACAGCAG------CAGCAACA------G------------------CAGCAG---------CAG-CA | |
| droEug1 | scf7180000409474:2542837-2543086 + | TTTG-------------------------------------------------------------TCATATTT-----GCAGTTAGCAACAACAACATTACAACAA----------------GTACAGC----------------T--AG-----------------------------------------------------T------------------------ACAGTTAGTGGAAACTACAACAACAACAACAACAACAAGCACC-----------------------------------------------------------------------------------TCAGTCACCA---AGCCCAAAACCAACCAACAAACTTTAAAAATAAAATCCACC--------------AAAAAAAACCAA-------------TCCA---------------------------------------------------ATCAACAAGAACAA-------------------------------------------------ACAACAACTACAAA-------AA---TAACAGCCACCTCCATTACAAGTACAACAACAACAACAA------CAACAACA------A------------------GAATAA---------CTA-GA | |
| dm3 | chr3R:27478328-27478539 + | AATTG--------------------------------------------------------TGTGCTGAGGTCATGTCGCTGGCGT---CGCCA--GTTGCAGCAG----------------CGGCAAC-------------------AA-----------------------------------------------------CAATGCAACAATGGGGCAGCAAC-------------------TGACGA---CT--ATAATGTCTGGC----------------------------CGGGCTCCACTCCTCTCACTCGCCTGGGCGGCATCAACTCAAATGGT--------------------------------------------------------------------------------------CTC-------------GCAGTTGTAAAGCTGC--------------------------------------------------------------------------------------------------------------------------------CAAGAGCA----------GACCGGCATGCGCAGCTGCAG------CAAACGCA------A------------------CAGCAG---------CAG-CA | |
| droSim2 | x:5444825-5445017 + | GCC------------------------------------------------------------AGGTGCCATC-----GCAGGTGGTCGCAGCA--GCCGCAGCAG----------------CTGCAGC-------------------GG-----------------------------------------------------C------------------------ACAGCAGCTGCAGG-TCCAGCATCAACAGCAGCAGCATCACC------------------------------------------------------------------------------------A-------------------------------------------------------------------TGTACAAACCAC-------------CCAGATTCAGCACATCCA---AGTGCAATC------------------------------------GGCACCG---CCCCCG--------------------------------------------------------------CAACAGCA---------GCAACAGCAACAGCAGCAGCAG------CAACAACA------G------------------CAGCAG---------CAG-CA | |
| droSec2 | scaffold_4:955813-956005 - | GCC------------------------------------------------------------AGGTGCCATC-----GCAGGTGGTCGCAGCA--GCCGCAGCAG----------------CTGCAGC-------------------GG-----------------------------------------------------C------------------------ACAGCAGCTGCAGG-TCCAGCATCAACAGCAGCAGCATCACC------------------------------------------------------------------------------------A-------------------------------------------------------------------TGTACAAACCAC-------------CCAGATTCAGCACATCCA---AGTGCAATC------------------------------------GGCACCG---CCCCCG--------------------------------------------------------------CAACAGCA---------GCAACAGCAACAGCAGCAGCAG------CAACAACA------A------------------CAGCAG---------CAG-CA | |
| droYak3 | X:14662695-14662887 + | GCC------------------------------------------------------------AGGTGCAATC-----GCAGGTGGTCGCAGCA--GCCGCAGCAG----------------CGG------------------------------------------------------------------------------C------------------------ACAGCAGCTGCAGG-TCCAGCAGCAACAGCAGCAGCATCACC------------------------------------------------------------------------------------A-------------------------------------------------------------------TGTACAAACCAC-------------CCAGATCCAGCACATTCA---AGTGCAATC------------------------------------GGCGCCG---CCCCCA------C--------------------------------------------------------AGCAGCA---------GCAGCAGCAACAACAACAGCAG------CAGCAACA------G------------CAATCACAGCAG---------CAA-CA | |
| droEre2 | scaffold_4929:5405373-5405543 - | CTGCC----------------------------------------------------------CGCCGCCCGT--------GGTG-ATGCGGAA--TCTGCAGCAG----------------CAGCAAC-------------------AA-----------------------------------------------------C----------------------A-ACATCAGCAG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCCAT-------------GCTCAGTC----------------------------------------C-----CAGCGGCAATGG----------------------------------CAATCTTTTGTATACTCAG-------------------------CA---ACAGCA------GCAGGGTCAGC------AGCAG------CAACCACAACAACAGCAGCAAGGACCACCAATTCGCTTT---------CAA-CA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/18/2015 at 02:09 AM