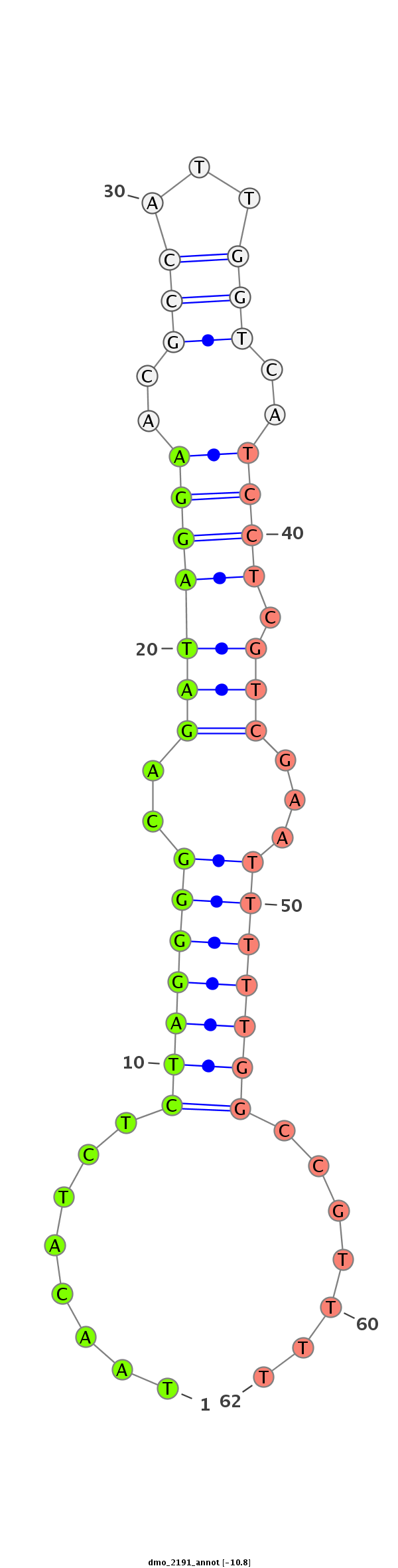
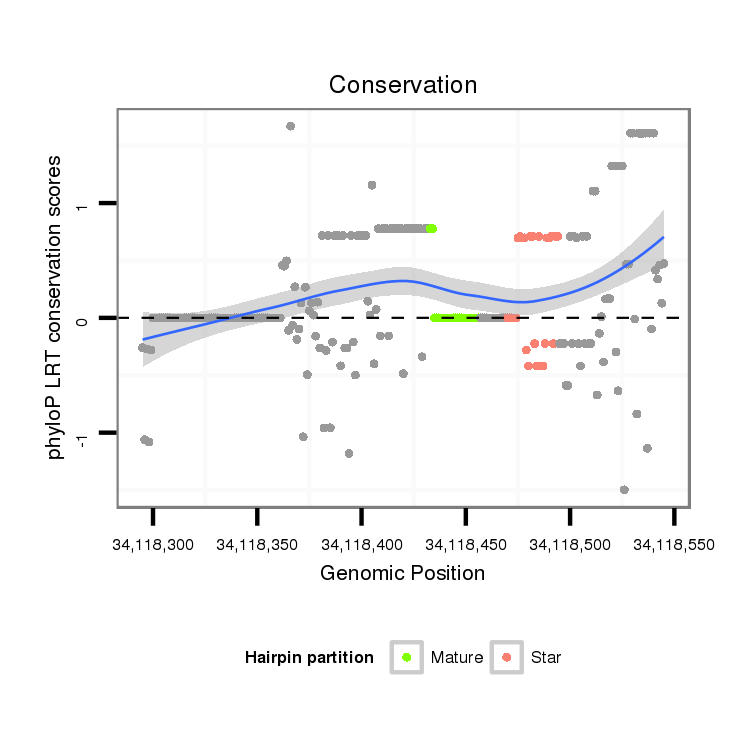
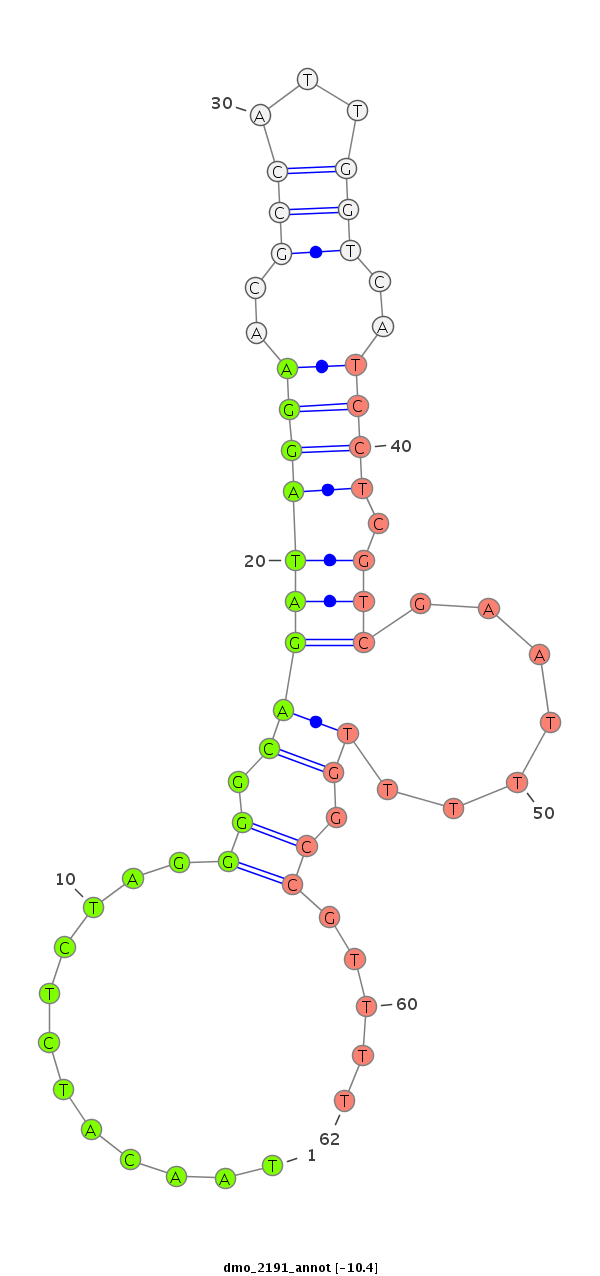
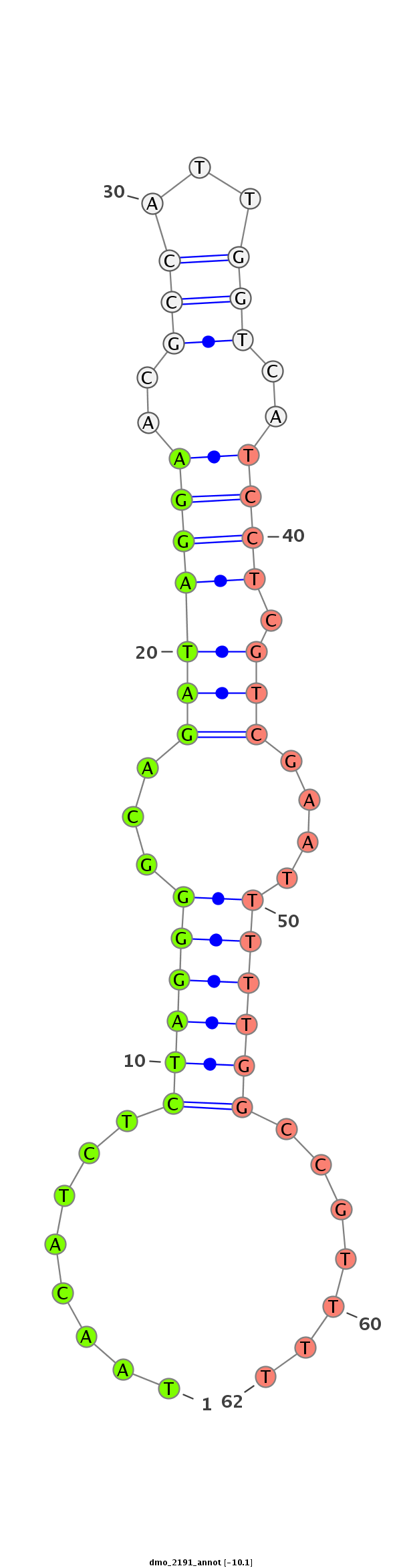
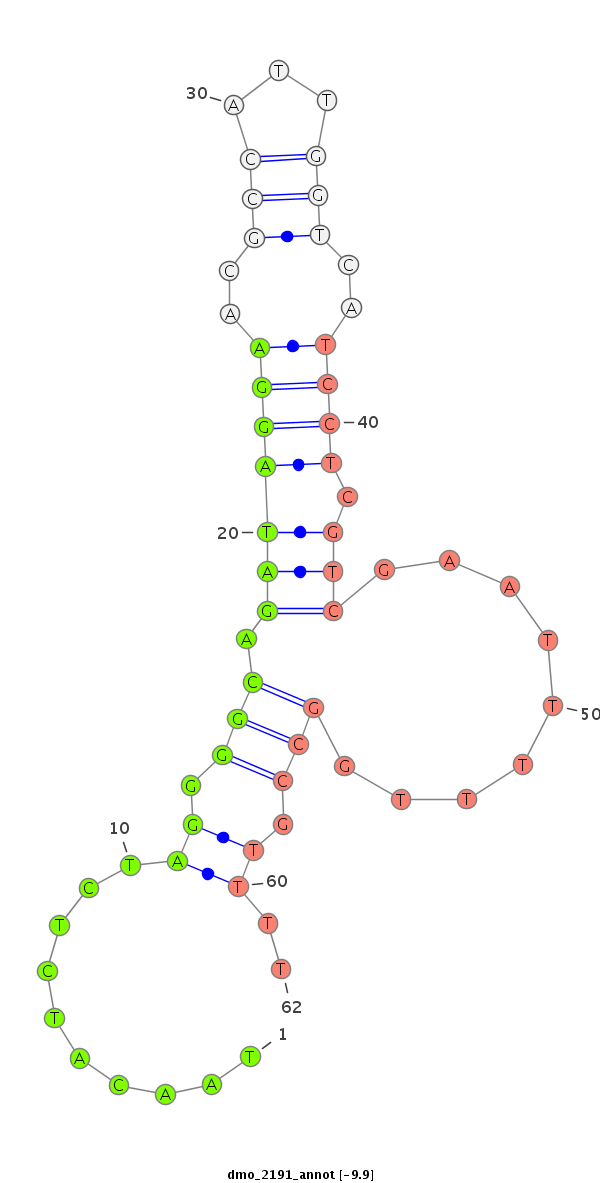
ID:dmo_2191 |
Coordinate:scaffold_6540:34118345-34118495 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -10.4 | -10.1 | -9.9 |
 |
 |
 |
exon [dmoj_GLEANR_10753:4]; CDS [Dmoj\GI10860-cds]; intron [Dmoj\GI10860-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AGATAGGAACGCCATTGGTCATCCTCGTCGAGTTTTTGGCCGTTTTGCAGGTAAGTGTATATTTTAAATTTTTGCTAGTTTTAATATTGTCATTAATTAGATGATTTTTTTAATTTTTCTCATTGCAGGTAAGCTTATTAACATCTCTAGGGGCAGATAGGAACGCCATTGGTCATCCTCGTCGAATTTTTGGCCGTTTTGCAGGTAAGTGTATATTTTAAATTTTTGCTAGATTTAATATTGTTATTAAT ******************************************************************************************************************************************........(((((((..(((((((..(((...)))..)))).)))...))))))).......*************************************************** |
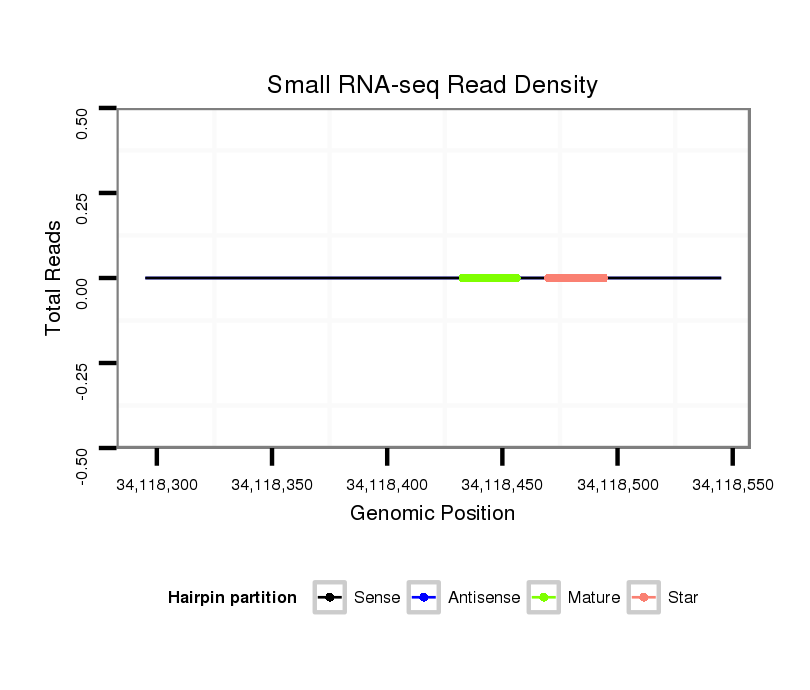
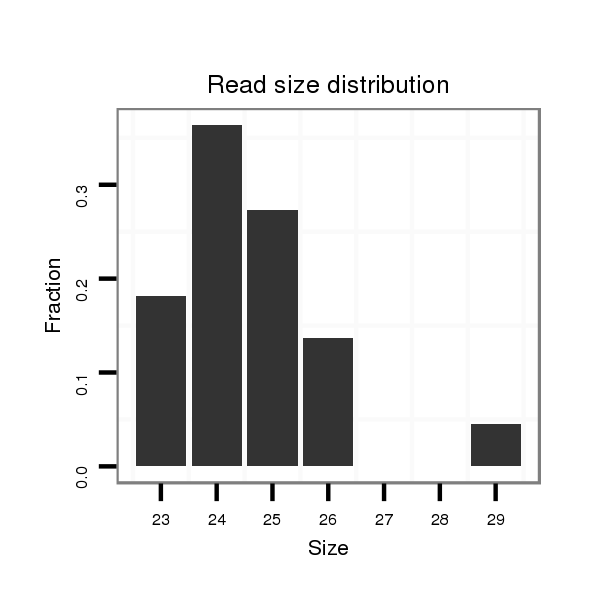
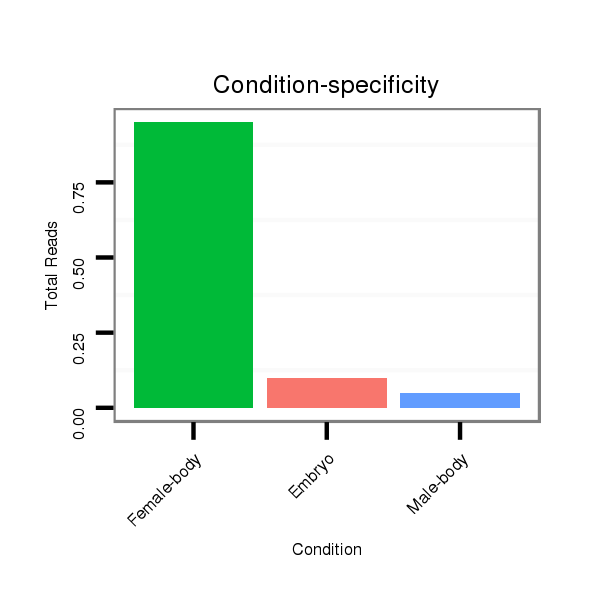
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V041 embryo |
V110 male body |
|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................TAACATCTCTAGGGGCAGATAGGA......................................................................................... | 24 | 0 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 |
| ...........CCATTGGTCATCCTCGTCGAGTTT........................................................................................................................................................................................................................ | 24 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................TAAGTGTATATTTTAAATTTTTGCTAGA.................. | 28 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ...............TGGTCATCCTCGTCGAGTTTTTGG.................................................................................................................................................................................................................... | 24 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 |
| ..............TTGGTCATCCTCGTCGAGTTTTTGGC................................................................................................................................................................................................................... | 26 | 0 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 |
| ...........................................................................................................................................AACATCTCTAGGGGCAGATAGGA......................................................................................... | 23 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 |
| ..........................................................................................................................................TAACATCTCTAGGGGCAGATAGGAAC....................................................................................... | 26 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ......................CCTCGTCGAGTTTTTGGCCGTTTTGC........................................................................................................................................................................................................... | 26 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| .GATAGGAACGCCATTGGTCATCCTCG................................................................................................................................................................................................................................ | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................TAACATCTCTAGGGGCAGATAGGAA........................................................................................ | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................TCCTCGTCGAATTTTTGGCCGTTTT................................................... | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ........................TCGTCGAGTTTTTGGCCGTTTTGC........................................................................................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................TTGCAGGTAAGCTTATTAACATCTCTAGG.................................................................................................... | 29 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................ATTAACATCTCTAGGGGCAGATAG........................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................AATTTTTCTCATTGCAGGTAAGCTT................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................AACATCTCTAGGGGCAGATAGGAAC....................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| .................................................................................................................................TAAGCTTATTAACATCTCTAGGGGC................................................................................................. | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ..........................................................................CTAGTTTTAATATTGTCATTAATT......................................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................TAACATCTCTAGGGGCAGATAGG.......................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................GATAGGAACGCCATTGGTCATCCTCG...................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................AAGCTTATTAACATCTCTAGGGGCA................................................................................................ | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................AGACCGGAACGCCGTTGG............................................................................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................TAAGTGTATATTTTAAATTTTTGCTAG................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................TAAGTGTATATTTTAAATTTTTGC...................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................GTAAGTGTATATTTTAAATTTT......................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .................GTCATCCTTGTCGAGTTTTTGGCC.................................................................................................................................................................................................................. | 24 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
TCTATCCTTGCGGTAACCAGTAGGAGCAGCTCAAAAACCGGCAAAACGTCCATTCACATATAAAATTTAAAAACGATCAAAATTATAACAGTAATTAATCTACTAAAAAAATTAAAAAGAGTAACGTCCATTCGAATAATTGTAGAGATCCCCGTCTATCCTTGCGGTAACCAGTAGGAGCAGCTTAAAAACCGGCAAAACGTCCATTCACATATAAAATTTAAAAACGATCTAAATTATAACAATAATTA
***************************************************........(((((((..(((((((..(((...)))..)))).)))...))))))).......****************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V041 embryo |
|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................................................TTAAAAACGATCTAAATTATA.......... | 21 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 |
| ..............................................................................................................................TCCATTCGAATAATTGTAGAGAT...................................................................................................... | 23 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 |
| .....................................................................................................................................................................GGTAACCAGTAGGAGCAGCTTAAAA............................................................. | 25 | 0 | 7 | 0.14 | 1 | 1 | 0 | 0 |
| ...................................................................................................................................................................GCGGTAACCAGTAGGAGCAGCTTATAC............................................................. | 27 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 |
| .......................................................................................................................................................CCGTCTATCCTTGCGGTAACCAGT............................................................................ | 24 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 |
| .......................................................................................................................................................CCGTCTATCCTTGCGGTAACCATT............................................................................ | 24 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ................................AAAAACCGGCAAAACGTCCATTCACATAT.............................................................................................................................................................................................. | 29 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ......................................................................................................................................................CCCGTCTATCCTTGCGGTAACCAG............................................................................. | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ......CTTGCGGTAACCAGTAGGAGCAGCT............................................................................................................................................................................................................................ | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ....................................................................................................................................................TCCCCGTCTATCCTTGCGGTAACCAGT............................................................................ | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .......................................................................................................................................................CCGTCTATCCTTGCGGTAACC............................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .................................................................................................................................................................TTGCGGTAACCAGTAGGAGCAGC................................................................... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6540:34118295-34118545 + | dmo_2191 | AG-ATAGGAACGCCATTGGTCATCCTCGTCGAGTTTTTGGCCGTTTTGCAGGTAAGTGTATATTTTAAAT------TTTTGCTAGTTTTAATATTGTCATTAATTAGATGATTTTTTTAATTTTTCTCATTGCAGGTAAGCTTATTAACATCTCTAGGGGCAGATAGGAACGCCATTGGTCATCCTCGTCGAATTTTTGGCCGTTTTGCAGGTAAGTGTATATTTTAAATTTTTGCTAGATTTAA----TATTGTTATTAAT |
| droVir3 | scaffold_12970:7515811-7515887 + | ATTATT--------------------------------------------------------------ATTATTATTATCATTATTATTAATTTAATTATTATTATTATTATTTTTATTA----------------------------------------------------------------------------------------------------------------------TTATTAT----TATTATTATTA-T | |
| droGri2 | scaffold_15074:7410880-7410953 - | TT-AAT--------------------------------------------------------------AT------TTTTGTTAATTTTGGTTTGGTAGTTAATATCACTATTTTTTTTA----------------------------------------------------------------------------------------------------------------------CATTTATTATTTATTTTTATTA-A | |
| droWil2 | scf2_1100000004762:1948323-1948410 - | AC-GCC--------------------------------------------------------------GT------CGTCGTCGTC-----------------------------------------------------------------------------------------------------GTCGTCTTCGTTTTCGTCTTATGCCTGAGCTTGTGCTTCTGCTCCTTAATAGATTTTG----TATTTTGATTGAT | |
| droBip1 | scf7180000391881:8034-8092 + | TTTGGT--------------------------------------------------------------AC------TTTTGAAGATAGAAGC-----------------------------------------------------------------------------------------------------------------------------------TTGAGACTTTTTTTAGATTTTG----TATTGTAATAAAT | |
| droKik1 | scf7180000302592:1991081-1991116 + | TCTTAT--------------------------------------------------------------A----------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTTTATATATTTAA----TATTTTTAAAAGT | |
| droEre2 | scaffold_4770:6275128-6275169 - | AA-ATA---------------------------------------------------------------T------TTTT-----------------------------------ATTCCTCTTTTTCATTGGAGGTAAGCGTATTA------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/18/2015 at 02:08 AM