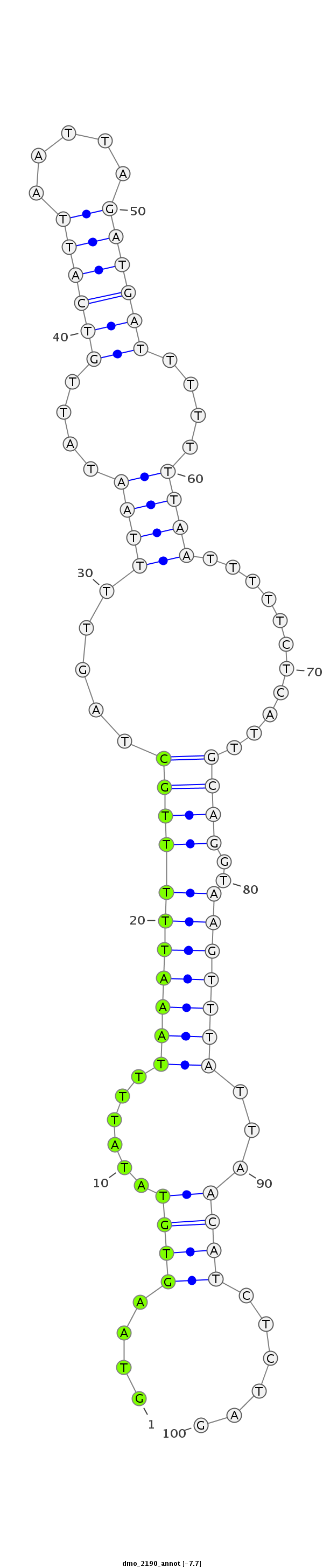

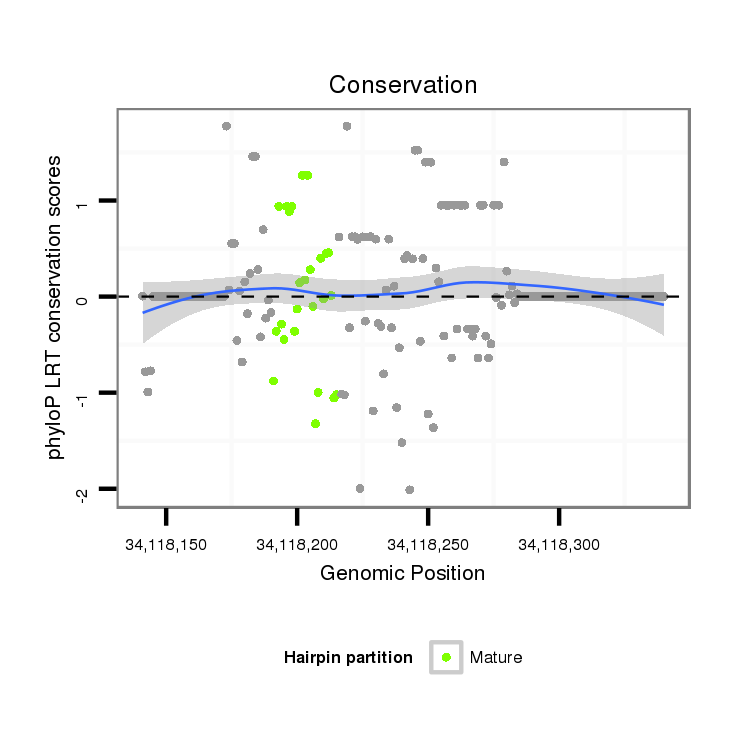
ID:dmo_2190 |
Coordinate:scaffold_6540:34118191-34118290 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -7.4 | -7.4 | -7.3 |
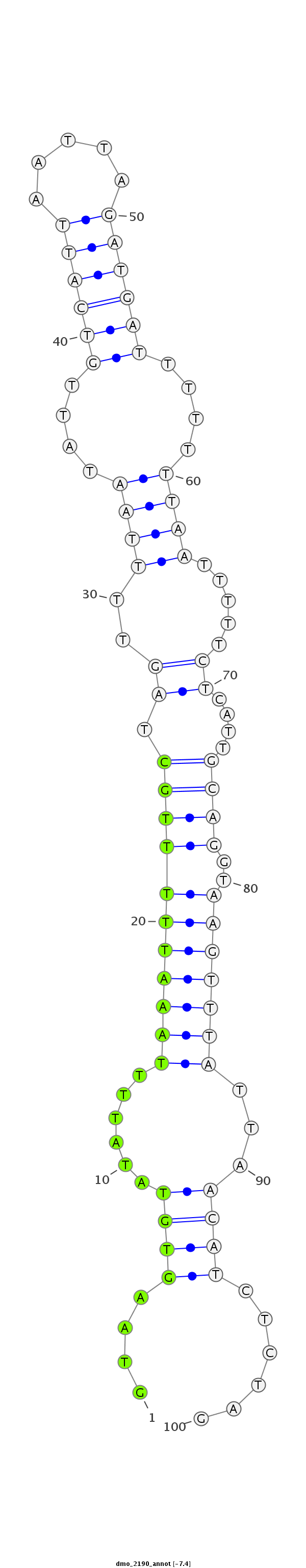 |
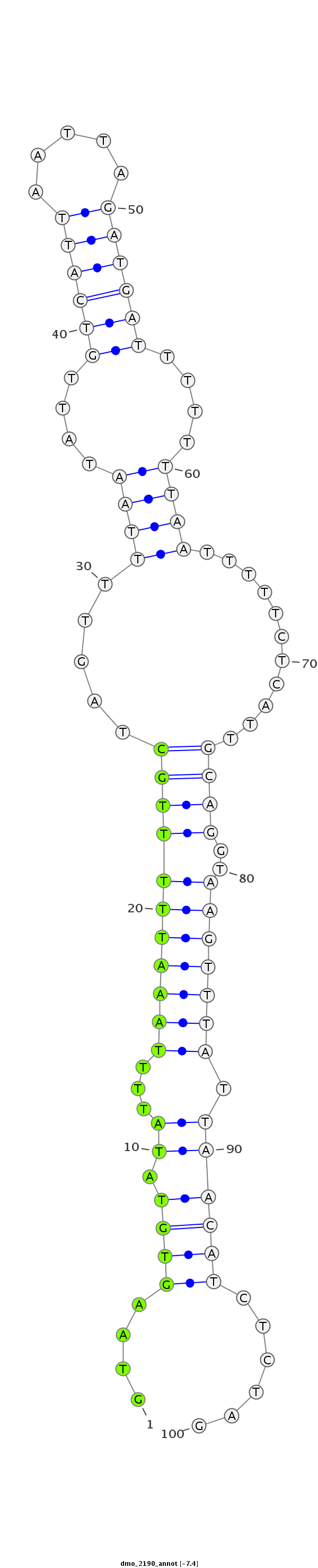 |
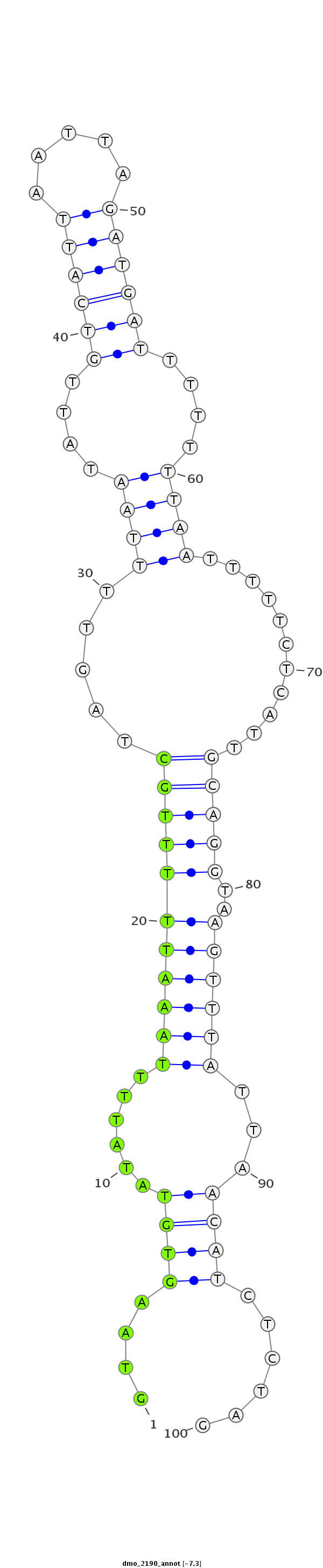 |
exon [dmoj_GLEANR_10753:3]; CDS [Dmoj\GI10860-cds]; CDS [Dmoj\GI10860-cds]; exon [dmoj_GLEANR_10753:4]; intron [Dmoj\GI10860-in]
No Repeatable elements found
| ##################################################----------------------------------------------------------------------------------------------------################################################## AGATAGAAGCGTCGTCAATCATCCTCGTCGAATTTTTGGCCGTTTTGCAGGTAAGTGTATATTTTAAATTTTTGCTAGTTTTAATATTGTCATTAATTAGATGATTTTTTTAATTTTTCTCATTGCAGGTAAGTTTATTAACATCTCTAGGGGCAGATAGGAACGCCATTGGTCATCCTCGTCGAGTTTTTGGCCGTTTT **************************************************....((((......(((((((((((.....((((....((((((.....))))))....))))...........))))..)))))))...))))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V041 embryo |
V110 male body |
|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................AACATCTCTAGGGGCAGATAGG....................................... | 22 | 0 | 20 | 0.40 | 8 | 8 | 0 | 0 | 0 |
| .....................................................................................................................................................................CCATTGGTCATCCTCGTCGAGTTT........... | 24 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 |
| .........................................................................................................................................................................TGGTCATCCTCGTCGAGTTTTTGG....... | 24 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 |
| ........................................................................................................................................................................TTGGTCATCCTCGTCGAGTTTTTGGC...... | 26 | 0 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 |
| ............CGTCAATCATCCTCGTCGAATTTT.................................................................................................................................................................... | 24 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ...........................................................................................................................................................GATAGGAACGCCATTGGTCATCCTCG................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..................................................GTAAGTGTATATTTTAAATTTTTGC............................................................................................................................. | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................TCCTCGTCGAATTTTTGGCCG.............................................................................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .....................TCCTCGTCGAATTTTTGGCCGTTT........................................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................GTAAGTTTATTAACATCTCTAGGGG............................................... | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...................CATCCTCGTCGAATTTTT................................................................................................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...................CATCCTCGTCGAATTTTTGGCCGTT............................................................................................................................................................ | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................GTCATCCTTGTCGAGTTTTTGGCC..... | 24 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................TCCTCGTCGAATTTTTGGCCGTTTTGC........................................................................................................................................................ | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ..........................................................................CTAGTTTTAATATTGTCATTAATT...................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
TCTATCTTCGCAGCAGTTAGTAGGAGCAGCTTAAAAACCGGCAAAACGTCCATTCACATATAAAATTTAAAAACGATCAAAATTATAACAGTAATTAATCTACTAAAAAAATTAAAAAGAGTAACGTCCATTCAAATAATTGTAGAGATCCCCGTCTATCCTTGCGGTAACCAGTAGGAGCAGCTCAAAAACCGGCAAAA
**************************************************....((((......(((((((((((.....((((....((((((.....))))))....))))...........))))..)))))))...))))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V041 embryo |
|---|---|---|---|---|---|---|---|---|
| .CTATCTTCGCAGCAGTTAGT................................................................................................................................................................................... | 20 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 |
| ..............................................................................AAAATTATAACAGTAATTAATCTACT................................................................................................ | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .......................................................................................................................................................CCGTCTATCCTTGCGGTAACC............................ | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .....CTTCGCAGCAGTTAGTAGGAGCA............................................................................................................................................................................ | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ......................................................................................................................GAGTAACGTCCATTCAAATAATTG.......................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ......................................................................................................................................................CCCGTCTATCCTTGCGGTAACCAG.......................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ................................................................................................................................................................CTTGCGGTAACCAGTAGGAGCAGCT............... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ..............................................................................................................................TCCATTCAAATAATTGTAGAGAT................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| .CTATCTTCGCAGCAGTTATT................................................................................................................................................................................... | 20 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ...............................................................................................................................CCATTCAAATAATTGTAGAGA.................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6540:34118141-34118340 + | dmo_2190 | AGATAGAAGCGTCGTCAATCATCCTCGTCGAATTTTTGGCCGTTTTGCAGGTAAGTGTATATTTTAAATTTTTGCTAGTTTTAATATTGTCATTAATTA-------------------------GATGATTTTTTTAATTTTTCTCATTGCAGGTAAGTTTATTAACATCTCTAGGGGCAGATAGGAACGCCATTGGTCATCCTCGTCGAGTTTTTGGCCGTTTT |
| droVir3 | scaffold_12970:7515712-7515788 + | ATTA----------------------------TT---A------------------------------------TTATTATTATTATTATTATTATTAA-------------------------TATTATTATTATTATTATTATTATTATTATTATTATTATTAATAT-------------------------------------------------------- | |
| droGri2 | scaffold_15252:1953517-1953604 + | CGAG----------------------------TTTTATACAGTTC---AATAAGTTGTTTATTTTAGTATCATACTTATATTAATATTTTCACTAAATA-------------------------GTTGATTAATTTGT-------------------------TCATTG-------------------------------------------------------- | |
| droWil2 | scf2_1100000004909:10466764-10466835 - | ATTT----------------------------TT---TGTTGTTTGGTTGCTAAGTGTAGGTTTGAGTTTTTTTTTTTTTTTTTTTTTGCCGTGGTT------------------------------------------------------------------TAATTG-------------------------------------------------------- | |
| droKik1 | scf7180000302469:1871490-1871586 + | ATTT----------------------------TA---TATTATTT---------------ATATTGTTTTTTATATTATTTTAATTTTTTTTTTAATTTTTTAATATTTTCTATATTTTTTAAAAATATTTTTTATAAT--------------------------TTTT-------------------------------------------------------- | |
| droFic1 | scf7180000454039:525437-525489 + | ATAT----------------------------TT---T---------------------------TCATGTTTGTCTATTAAAGCACTTTCACTAATTG-------------------------GAAGATTTT---------------------------------CTT-------------------------------------------------------- | |
| droEle1 | scf7180000491349:25084-25172 + | TATT----------------------------TA---TTTTATTT------------------------ATTTATTATTATTATTATTATTATTATTAT-------------------------TATTATTATTATTATTATTATTATTATTATTATTTTAAGTAATAT-------------------------------------------------------- | |
| droRho1 | scf7180000777514:2661-2721 + | TAAA----------------------------TT---T--------------------------TAAGTTTTCAATCATTTTACTTTAATTTGAAAACA-------------------------AATCATTTTTCTAAC--------------------------TTTT-------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
Generated: 05/18/2015 at 02:06 AM