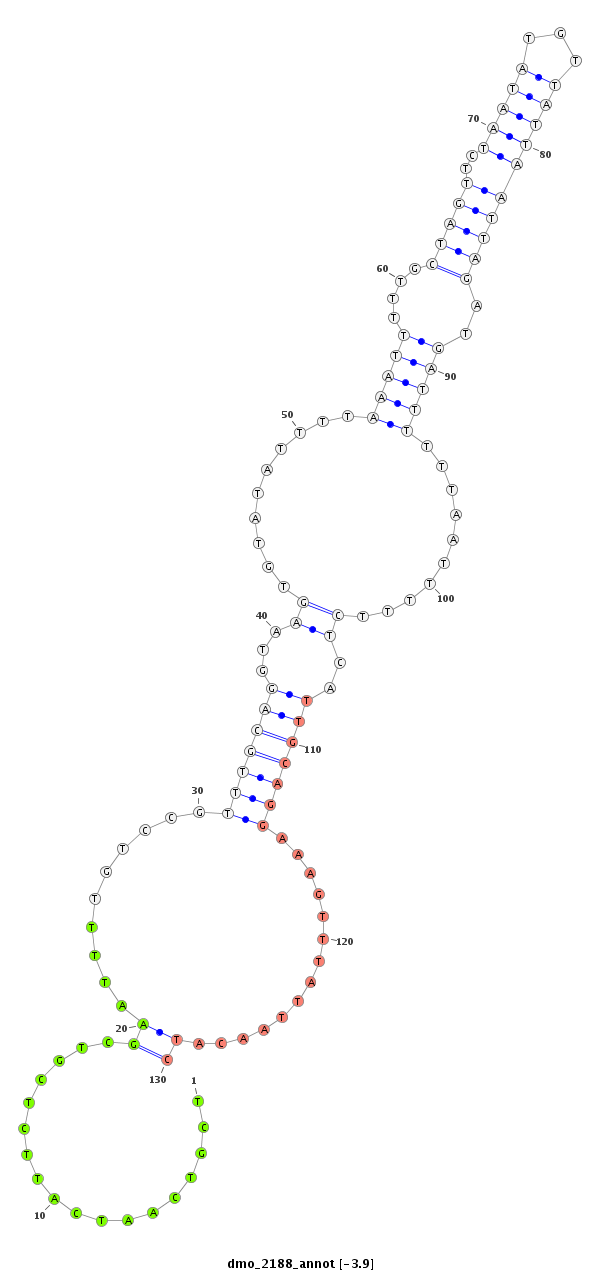
| ..............................................................................................................................................................................................AACATCTCTAGGGGCAGATAGGA...................................... |
23 |
0 |
20 |
0.15 |
3 |
3 |
0 |
| ...........................................................................................................................................................................................................................TTGGTCATCCTCGTCGAGTTTTTGGC...... |
26 |
0 |
20 |
0.15 |
3 |
0 |
3 |
| ............................................................................................................................................................................................................................TGGTCATCCTCGTCGAGTTTTTGG....... |
24 |
0 |
20 |
0.15 |
3 |
3 |
0 |
| ........................................................................................................................................................................................................................CCATTGGTCATCCTCGTCGAGTTTTT......... |
26 |
0 |
20 |
0.10 |
2 |
2 |
0 |
| ..................................................................TCGTCAATCATTCTCGTCGAATTT................................................................................................................................................................. |
24 |
0 |
20 |
0.10 |
2 |
2 |
0 |
| ..................................................................TCGTCAATCATTCTCGTCGAATTTT................................................................................................................................................................ |
25 |
0 |
20 |
0.10 |
2 |
1 |
1 |
| ...............................................................................TCGGCGACTTTCGTCCGTTT........................................................................................................................................................ |
20 |
3 |
10 |
0.10 |
1 |
1 |
0 |
| ..................................................................TCGTCAATCATTCTCGTCGAAT................................................................................................................................................................... |
22 |
0 |
20 |
0.05 |
1 |
0 |
1 |
| ................................TAAGCTTATTAACACTCTAGGGG.................................................................................................................................................................................................... |
23 |
0 |
20 |
0.05 |
1 |
1 |
0 |
| .....................................TTATTAACACTCTAGGGGC................................................................................................................................................................................................... |
19 |
0 |
20 |
0.05 |
1 |
0 |
1 |
| .....................................TTATTAACACTCTAGGGGCAGAT............................................................................................................................................................................................... |
23 |
0 |
20 |
0.05 |
1 |
1 |
0 |
| .................................AAGCTTATTAACACTCTAGGGG.................................................................................................................................................................................................... |
22 |
0 |
20 |
0.05 |
1 |
1 |
0 |
| .............................................................................................................................................................................TTGCAGGAAAGTTTATTAACATC....................................................... |
23 |
0 |
20 |
0.05 |
1 |
1 |
0 |
| .............................................................................................................................................................................................TAACATCTCTAGGGGCAGATC......................................... |
21 |
1 |
20 |
0.05 |
1 |
1 |
0 |
| .................................................................CTCGTCAATCATTCTCGTCGAATT.................................................................................................................................................................. |
24 |
1 |
20 |
0.05 |
1 |
0 |
1 |
| .................................AAGCTTATTAACACTCTAGGGGCAGAT............................................................................................................................................................................................... |
27 |
0 |
20 |
0.05 |
1 |
1 |
0 |
| ..............................................................................................................................................................................................................................GTCATCCTTGTCGAGTTTTTGGCC..... |
24 |
1 |
20 |
0.05 |
1 |
1 |
0 |
| ...........................................................................................................................................................................CATTGCAGGAAAGTTTATTAACATCTC..................................................... |
27 |
0 |
20 |
0.05 |
1 |
1 |
0 |
| .........................................TAACACTCTAGGGGCAGATAGA............................................................................................................................................................................................ |
22 |
0 |
20 |
0.05 |
1 |
1 |
0 |
| ........................................................................................................................................................................................TTTATTAACATCTCTAGGGGC.............................................. |
21 |
0 |
20 |
0.05 |
1 |
1 |
0 |
| ....................................CTTATTAACACTCTAGGGGCAGATA.............................................................................................................................................................................................. |
25 |
0 |
20 |
0.05 |
1 |
0 |
1 |
| ................................TAAGCTTATTAACACTCTAGGGGCATT................................................................................................................................................................................................ |
27 |
2 |
20 |
0.05 |
1 |
1 |
0 |