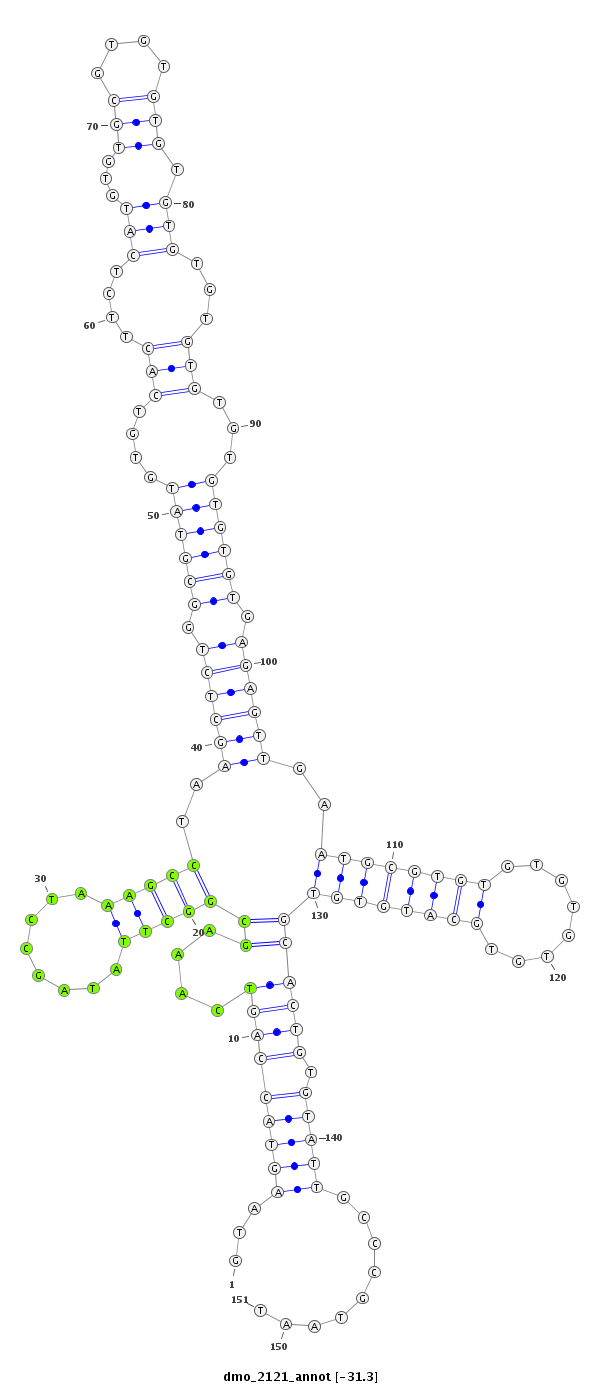
| droMoj3 |
scaffold_6540:29769001-29769251 + |
dmo_2121 |
CT---------------------GTCTGCATGGCCAAAATGTCGGCAGCCGGTTCCCCAGAGGAGGAGGTTGTAAGTACCAGTC-----------------------------------------------------------------------------A----------------AAGCGGCTTATAGCCTAAAGCCTAAGCTCTGGCGTATGTGTC--------ACTTC-TCA--TGT----------------------GTGCGTGTGTGTG------T-------------------GT---------------------G-TGTGTGTGTGTGT--------------------GAGAGTTGA--AT--------G----CGT------------GTGTG--------T--GTGTGCATGTG-----------------------------------------------------------------T--------------GCACTGTGTATTGCCCGT---------AATTGGCGCCAATTTCACTGTGGCCA-------GTTGAATATTTTGGCG---------------------------ACTCATAAGCA |
| droVir3 |
scaffold_13047:12012022-12012216 - |
|
GT---------------------GCCTGCATGGCCAAAATGTCGGCAGCTGGTGCGCCCGAGGAGGAGCAGGTAAGTACCAGCC-----------------------------------------------------------------------------A----------------AAGCGGCTTATAAGCTTAAGCCTCAGCTCAAGCGTATGTGTT--------GCTG-------TTC---GT------GAG---T------------------------------------------------------------------G---T-------------------------------------------GCGTGTGTGTGC--------------G------CG-------------------------------------------------------------------------------------------------------CTGTACTTTGTTCGT---------AATTGGCACCAATTTCA----------------GTTGAATATTTTTGCG---------------------------ACTCATAAGCA |
| droGri2 |
scaffold_14624:3498436-3498604 - |
|
AA---------------------GG--------------------------------------ATCAGTCAATAATGCTG-----------ACAT--CAAATTGATGCCAATCTGACGGCTCTGTCATCTCA-------------------------------------------------------------------------------------------------CAAT-TCA--TGTG------------------A--G--TGTGTGTGTGT----GTGTGTGTATG--TATGTGT-GTGT-------------------------------------------------------------------------TTGTGTGTGTGTGAG----TGTGT--T--------T--GTCTGT----T--------------------------TTTATGCGGGTT------------------------GTGTGCGT-------------------------------------TTTT-----CAATTTCA--------------------------------------------------------------------CT |
| droWil2 |
scf2_1100000004943:2637853-2638139 + |
|
GT---------------------GCTTGTATGGCCAAAGTGACGTCGTCTGGTGCCCCCGAGGAGGAGCATGTAAGTACCCGTTTGTCCCTACAACAATTTCTAAAACATAGTGGGAC----------------------GAGAAATGAGGGGAGTAGTTCTTAGTTCACCTAAAAGAAAGCCGCTTATGGGCTT----------------------AGAGCTCTTTG-----------CATCTA-TGGTAG-----------------TGTGTGCC--------TCTGTCTCTG--TGTGT-GTGT------------GTGTGTT----------TGTGTGGGAACGTAATTTTCTGGACC--------------------------------------------------------C------------------------------------------------------------------------------------------------------------TCTGCTCCTTTTCCTTTCCATTCTCATCCTCGTC--------GTTGAAGTCATCG-------------------------------------TCGA |
| dp5 |
2:4789060-4789293 + |
|
GT---------------------GCCTGCATGGCCAAAGTGGCGGCCTCCGGTGCCCCCGAGGAGGAGCATGTAAGTACC-----------ACAG--AAGGCTAACACAAAGCGAGAATGTTGGCCACCTGAATGAAGCAGCTAAAGAAGGAAAG------------------AAAGAAAGCCGCTTATGTGCCT----------------------AAAACTCTTTGCC---C---A-GCC---AT------GCA---T------------------------------------------------------------------C---T-------------------------------------------ACGTATCCGTGT--------------G------CG----------------------------------------------------------G------------------------TCT--------------ACCCTGCGGCTCGGTCGT---------AAATTGCAC-----------------------------------------CTCCTTTG------------------GATCGAAAACG |
| droPer2 |
scaffold_19:485857-486090 + |
|
GT---------------------GCCTGCATGGCCAAAGTGGCGGCCTCCGGTGCCCCCGAGGAGGAGCATGTAAGTACC-----------ACAG--AAGGCTAACACAAAGCGAGAATGTCGGCCACGTGAATGAAGCAGCTAAAGGAGGAAAG------------------AAAGAAAGCCGCTTATGTGCCT----------------------AAAACTCTTTGCC---C---A-GCC---AT------GCA---T------------------------------------------------------------------C---T-------------------------------------------ACGTATCCGTGT--------------G------CG----------------------------------------------------------G------------------------TCT--------------ACTCTGGGGTTCGGTCGT---------AAATTGCAC-----------------------------------------CTCCTTTG------------------GATCGAAAACG |
| droAna3 |
scaffold_12929:1384606-1384692 - |
|
TC---------------------TCATGTGT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------G--TGTGTGTGTGT----GAGTGTGTGTG--TGTGTGT-GTGA-------------------------------------------------------------------------GTGTGTGTGTGTGCCCAAGTT---------------------------------------------------------------------------------------------------------------TTGTGTATTTTTCGG---------GAATGGC-------------------------------------------------------------------------------- |
| droBip1 |
scf7180000396712:1708826-1708903 - |
|
GT---------------------GTATGTGTATG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T--GTATGTGTATGTG-------------------------TAT---------------------G-TGTATGTGTATGT--------------------GTATGGTGT--AT--------G----TGT------------ATGTG--------T--ATGTGTATATG-----------------------------------------------------------------T--------------G----------------------------------------------------------------------------------------------------------------- |
| droKik1 |
scf7180000299277:117385-117466 - |
|
CTGT----------CA--------TGTGTGTGC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAG---------------C---GT--GT--G----TGTGTGTG--------TGTGTGTG--TGTGT---GTGT------------------------GTGTGTGT---------------------------GT--------------G----TGTGT--G------TGTGTG--------T--GTGTGC----T-----------------------------------------------------------------T--------------G----------------------------------------------------------------------------------------------------------------- |
| droFic1 |
scf7180000453882:768504-768645 + |
|
TG----------------------TG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T--GT--GTGTGTGTG--------------TG---------TGT---------------------G-AGTTTGCGTGTGT--------------------GATTCGAGA------------------GTGT--G------TCTGTG--------T--GTGTGT--CTGACAGCTGTCAAGGTGTTGACATC---GG----------------------------------------------AAATTG------------------CGTAGGAAA-------------------------------------------------TTTCAAAAGTGCCTGAC-ATTTCGGGCATTTTTGAGCG |
| droEle1 |
scf7180000491006:765157-765290 - |
|
CT---------------------TCGCATGT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------G--TGTGTGTGTGT----GTGTGTGTGTG--AGTGTGT-GTGT-------------------------------------------------------------------------GTGTGTAGGTGT---CAAGGG---------------------------T-----------------GCTGTCCTGGCC----------GGGAAACGAAAGCCCCACCACCA--------CCCT------------GGGCGGGA-----------------------------------------------TTGCATATTTTGCTG---------------------------CCTCA--CGAA |
| droRho1 |
scf7180000779876:153657-153808 + |
|
AC---------------------GTGG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A--------ACTTT-TCA--CACATG-CAGCAGT---GT--GT--G----TGTGTGTG--------------TG---------TGCGAGA---GGTTTGAGTGTGTT----------AGT---------------------------GCG------------------AGTGT--G------CATATG--------T--ATGTGT----T--TACAGTTA-------------GCGGGC----------GCCAAACCAAG--------------------------------------------------------------------------G--------TGTCGCC----------------GTGCACCCAAAAT----CGTTT-----CAGCGTGAATA |
| droBia1 |
scf7180000302193:855944-856043 - |
|
GCTG--CGGTGTGT---------------GTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTT-CTG--TGTGTG-TGTGTGTGT-GT--G--------TGTGTGTGTGTGTGTGTGTGT--GTGT--GT---------------------GTGTG-T-G------T-----------------------------GT--------------G----TGTGT----------GT--GTGTGTGTGTGTGTGT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droTak1 |
scf7180000414555:16948-17007 + |
|
CG----------------------TG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T--GT--GTGTGTGTG--------------TG---------TGT---------------------G-TGTGTGTGTGTGT--------------------GTGTGGTGA------------------GTGC--T------CGAGTG--------T--GTGT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droEug1 |
scf7180000409061:257736-257864 + |
|
TGCT----------ACTGTTCGTGTG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T--G--TGTGTGTGTG--------TGTGTGTGTG--TGTGT-GTGT------------GTGTGTT----------AGTGTG--------------------GTACGGCA------------------GTGT--G------TGTGTG--------T--GCGTGC----T--------------------------------------------------------------GAGTAGGCTCTTGAGCCG------------------CACAACACT-------------------------------------------------TCCGGTTG-----------------------CCACTTG |
| dm3 |
chr3L:10799841-10800041 - |
|
TG----------------------TGTGTGTGCCCAAAAGG-CATTTCCCGGCGC-----------AGT--GAGAAAATCAATC-----------------------------------------------------------------------------AAAA---------------TCGATTTAAAAGCACAACTTCCG----TTGCGGATTTACC--------AATGT-CCG--TGTGT--------------------G--CCCGGGTGTG------T------GCG--TGTGTGT-GTGGGTGAATGTTTGCCATTGGG----------CT---------------------------TGTG------------------GGTGT--A------TGTATG--------G--GTGTGT----G--TATTGTGA-------------------------------------------------------------------------------------------------AAATGTTACCAATTTATT--------TG----------------------------------------------------------T |
| droSim2 |
3r:14127107-14127223 - |
|
GC---------------------TTGTGCGTAAG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T---GT--GA--G----TGTGTGTG--TGTGTGTGTGT--GTGT--GTGT-GT---------------------GT-G------T-------------------------------------------GTG----TGTGT----------GT--GTGGACGTATGTGAGT------T-----------------GGCATA---GA----------------------------------------------AAATTA------------------TTTGCTACT-------------------------------------------------TTTAATTA-----------------------GTGCCAA |
| droSec2 |
scaffold_24:231369-231484 - |
|
AGCGCACGGCATAT---------------GTA--------------------------------------------------------------------------------------------------------------------------------------------------------CTCACGTATGT----------------------ATTGTCTCCT-----C-CCC--TGC---AT------GTA-TATGTGTGT--GTGTGTGTG--------------TG---------AGGGT----------------------GAGGGTGAGGGT--------------------GTTTTGCGG------------------GTGT--G------TGCGTG--------T--GTATCC----T--------------------------------------------------------------------------------G----------------------------------------------------------------------------------------------------------------- |
| droYak3 |
X:6555407-6555550 + |
|
GCAC--CGCCATAC-----------GTGTTTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C---------------TGCCGTTT---AC------ATG-----T--GTGTGTGTGTGTGT-------------------------GA---------------------G-TGTGTGTGTGCGT--------------------GAGGGGGTTAAAT--------------GC------------GAGTG----CATGT--AAAT----GTT-----------------GGCGCCGTGGCCAA-------------ACAATA-------------------------------------------------------------GCCGCTA--------------------------GTTAGTTTGCTG---------------------------GCCA--GTTGA |
| droEre2 |
scaffold_4690:9760258-9760417 - |
|
CT---------------------CTGTCGGTGGC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCTGC-CTC--TGTGT-GC---------------------GTGCGTGCG------A--------GTGT--GTGT-CT---------------------GT-GTGTGTCGTTCTG--------------------TTGCTGTTGTGT--------------GT------------GTGCG----TGCGT--GTGC----ATT-----------------GCCACTGTGTGCGTGCGAGTAG------------------------TGC--------------CTACTGCGGCCGAGTCGA---------GTGTTGCCT-----------------------------------------GTGCTTC-----------------------AAAATCG |