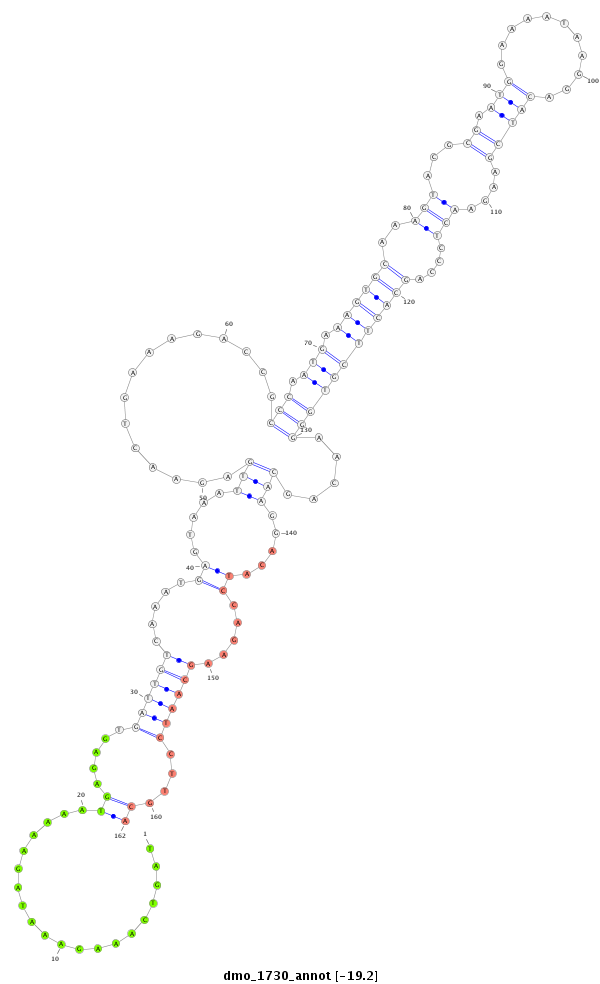
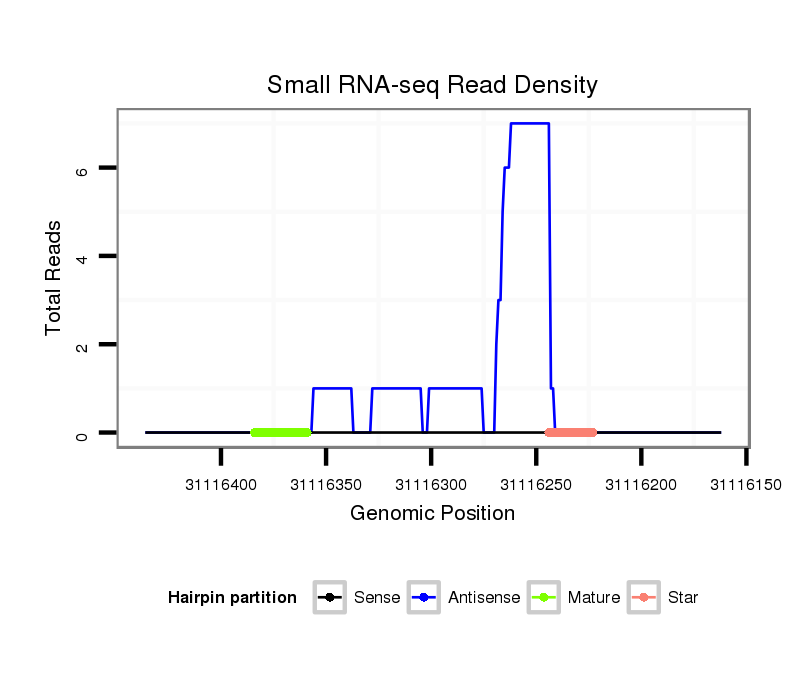
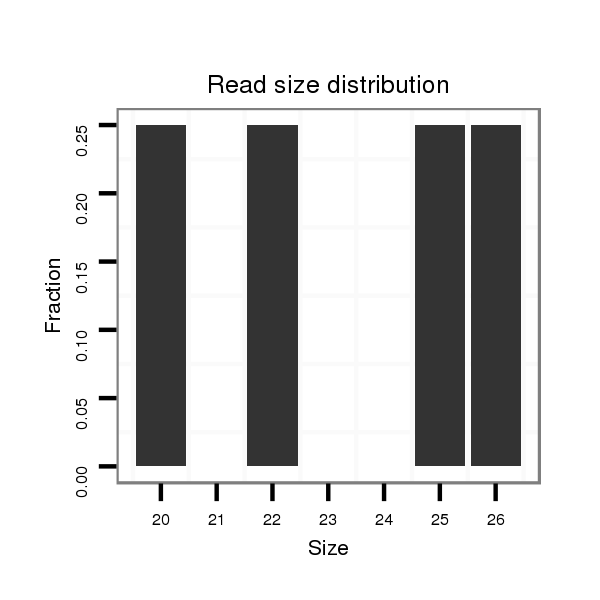
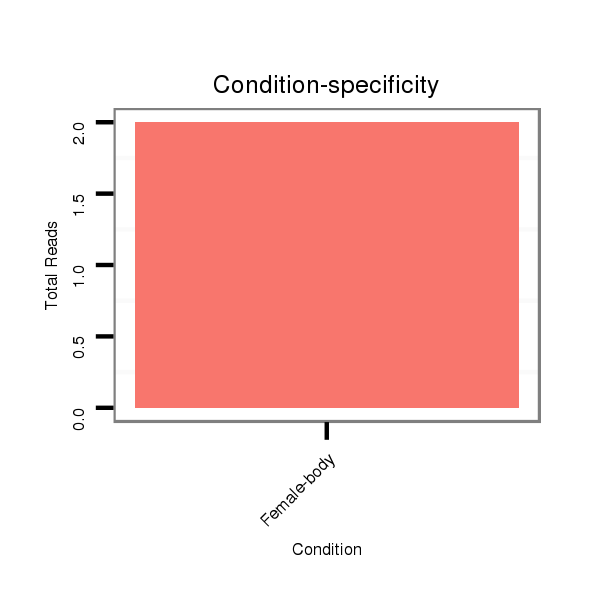
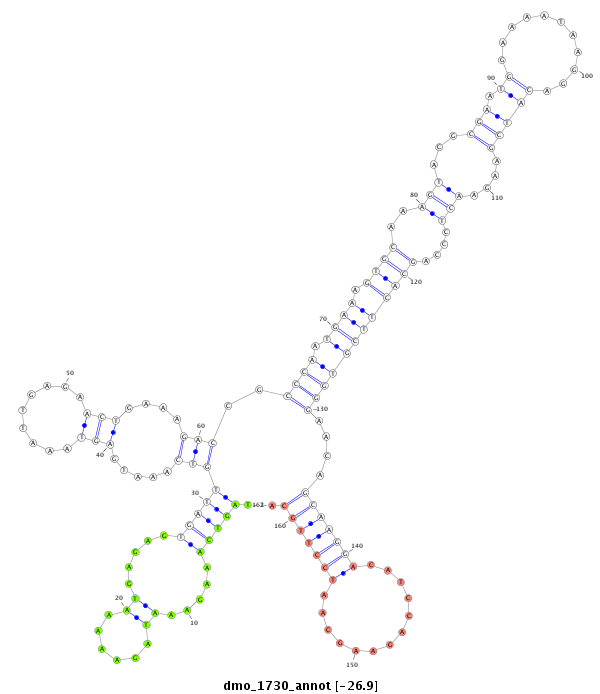
ID:dmo_1730 |
Coordinate:scaffold_6500:31116212-31116386 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -26.9 | -26.9 | -26.9 | -26.9 |
 |
 |
 |
 |
exon [dmoj_GLEANR_381:3]; CDS [Dmoj\GI18873-cds]; exon [dmoj_GLEANR_381:2]; CDS [Dmoj\GI18873-cds]; intron [Dmoj\GI18873-in]
| Name | Class | Family | Strand |
| OSVALDO_LTR | LTR | Gypsy | - |
| mature | star |
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AAGCAGGGCCTGTGGCTGGGCTATAAGAGGCCGCTCGCAGCTCCATACGGGTTAGTCAAAGAAATAGAAAAATGAGAGTGATTGTCAAATGAGTAAATTGAGAACTGAAAGACCGCCCAATGAAAGTGCAAAGTACGCGAATGGAAAATAAGGACATCGAAGAACTCCCAGCACTTCGTGGGAACAGCAAGGACATCCAGAAGCAATCCTTGCAACAACTTTCAGAGGAGAAGGGGCAAGCAGGAACGTCAGCGGCACCAACGAGGACATTGATG ****************************************************....................((.....((((((.....((.....(((...............(((.(((.((((((..(((...((.(((...........)))))....)))....)))))))))))).....))).....)).....))))))....))************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V056 head |
|---|---|---|---|---|---|---|---|
| ....................................................TAGTCAAAGAAATAGAAAAATGAGAG..................................................................................................................................................................................................... | 26 | 0 | 2 | 0.50 | 1 | 1 | 0 |
| ................................................................................................................................................................................................ACATCCAGAAGCAATCCTTGCA............................................................. | 22 | 0 | 2 | 0.50 | 1 | 1 | 0 |
| .......................................................................................................................................................................................ACAGCAAGGACATCCAGAAG........................................................................ | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 |
| ................................................................................................................................................................................................ACATCCAGAAGCAATCCTTGCAACA.......................................................... | 25 | 0 | 2 | 0.50 | 1 | 1 | 0 |
| ...........GTAGCTGGGCTATAAGAGC..................................................................................................................................................................................................................................................... | 19 | 2 | 9 | 0.11 | 1 | 0 | 1 |
| ...........................................CATACGGGTTAGATAACGA..................................................................................................................................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 1 |
| ..GCAGGGCCTGTGGCTGGGCTA............................................................................................................................................................................................................................................................ | 21 | 0 | 14 | 0.07 | 1 | 0 | 1 |
|
TTCGTCCCGGACACCGACCCGATATTCTCCGGCGAGCGTCGAGGTATGCCCAATCAGTTTCTTTATCTTTTTACTCTCACTAACAGTTTACTCATTTAACTCTTGACTTTCTGGCGGGTTACTTTCACGTTTCATGCGCTTACCTTTTATTCCTGTAGCTTCTTGAGGGTCGTGAAGCACCCTTGTCGTTCCTGTAGGTCTTCGTTAGGAACGTTGTTGAAAGTCTCCTCTTCCCCGTTCGTCCTTGCAGTCGCCGTGGTTGCTCCTGTAACTAC
*************************************************************....................((.....((((((.....((.....(((...............(((.(((.((((((..(((...((.(((...........)))))....)))....)))))))))))).....))).....)).....))))))....))**************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V041 embryo |
V110 male body |
V049 head |
V056 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................CGTGAAGCACCCTTGTCGTTCCT.................................................................................. | 23 | 0 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................GGTCGTGAAGCACCCTTGTCGTTCCT.................................................................................. | 26 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................CACCCTTGTCGTTCCTGTAGGTCATCGT...................................................................... | 28 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................GTGAAGCACCCTTGTCGTTCCTGT................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................GCGCTTACCTTTTATTCCTGTAGCTT.................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................TTCTGGCGGGTTACTTTCACGTTT............................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................GTCGTGAAGCACCCTTGTCGTTCCT.................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................TAACAGTTTACTCATTTAA................................................................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................AAGCACCCTTGTCGTTCCT.................................................................................. | 19 | 0 | 3 | 1.00 | 3 | 2 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................CGTTAGGAACGTTGTTGAAAGTCT................................................. | 24 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................................................TTCGTTAGGAACGTTGTTGAAAGTCT................................................. | 26 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................CCGTTCGTCCTTGCAGTCGCCGTGGT............... | 26 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................CTTCGTTAGGAACGTTGTTGAAAGTCT................................................. | 27 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................CCTTGCAGTCGCCGTGGTTGCTCCTG....... | 26 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................AGCACCCTTGTCGTTCCTGTAGGT............................................................................ | 24 | 0 | 20 | 0.30 | 6 | 0 | 6 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................TGGGTATTCGTTAGGACCGT............................................................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................CTTGTAGCTTCTCGAGGGT......................................................................................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................................................................TTGTTGAAAGTCTCCGCCTTC......................................... | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................AAAGCACCCTTGTCGTTCCT.................................................................................. | 20 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................AAAAGCACCCTTGTCGTTCCT.................................................................................. | 21 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................AAAAAGCACCCTTGTCGTTCCT.................................................................................. | 22 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................TGCAGTCGCCGTGGTTGTTCC......... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................AGCACCCTTGTCGTTCCTGTAGGTC........................................................................... | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................CCCTTGTCGTTCCTGTAGTT............................................................................ | 20 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................GCACCCTTGTCGTTCCTGTAGGTCT.......................................................................... | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................CCCTTGTCGTTCCTGTAGGTCT.......................................................................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................CACCCTTGTCGTTCCTGCTAG............................................................................. | 21 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6500:31116162-31116436 - | dmo_1730 | AAGCAGGGCCTGTGGCTGGGCTATAAGAGGCCGCTCGCAGCTCCATACGGGTTAGTCAAAGAAATAGAAAAATGAGAGTGATTGTCAAATGAGTAAATTGAGAACTGAAAGACCGCCCAATGAAAGTGCAAAGTACGCGAATGGAAAATAAGGACATCGAAGAACTCCCAGCACTTCGTGGGAACAGCAAGGACATCCAGAAGCAATCCTTGCAACAACTTTCAGAGGAGAAGGGGCAAGCAGGAACGTCAGCGGCACCAACGAGGACATTGATG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
Generated: 05/18/2015 at 01:10 AM