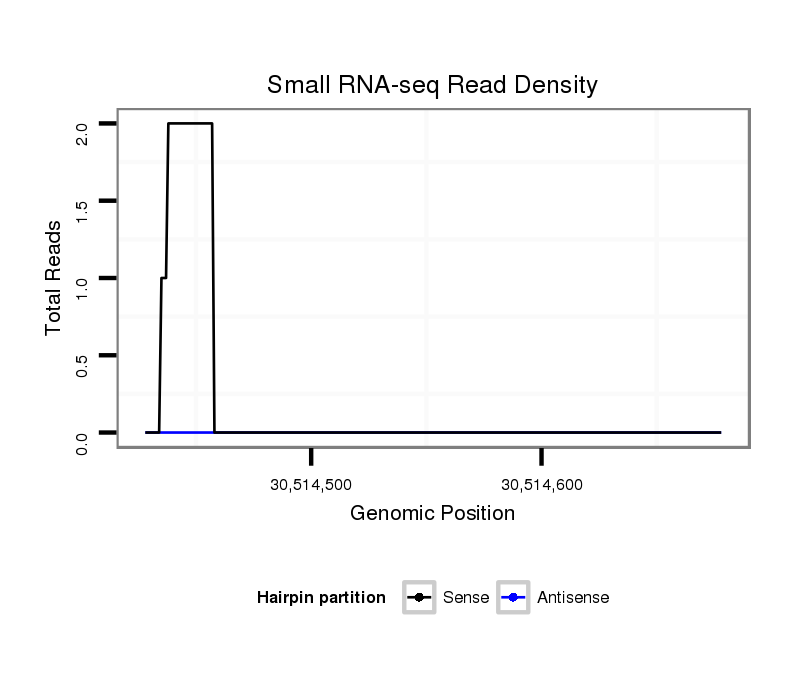
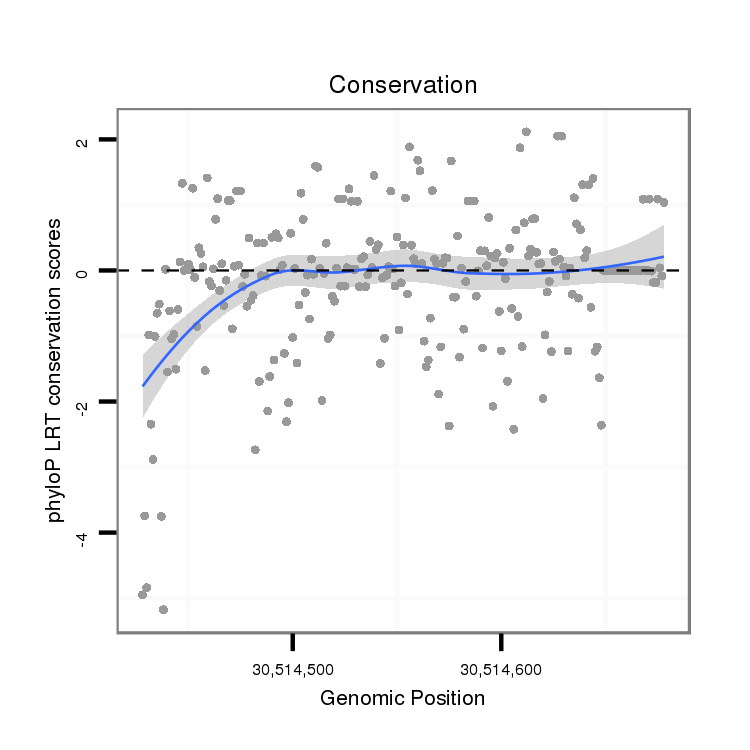
| droMoj3 |
scaffold_6500:30514428-30514678 + |
dmo_1719 |
CATTGTTTTTTTATTGGGAACATTTTTGTTTACGAAAGACTTT-----ATAAATGGTG-------------------AGTGGTTAGTGATTATTGAAACCATGCAG-----------------------------------AT--------------------------------------------------T-ATTA----------------------------------------------------------------------------AGAAGATAAATATTAA-------------AATAAT------------------------------------------------------AT-------------------------------------------------------------------------------------------ATTATA---AAT--------------------------------------------AAA----------------------TT---------------TAAATAATTCTTAAAGTAAAG--AAAAG-----------------------------------------------------------------------------------------AGAAAAAGTAAAAAAAAAGTAAGAAA-------------------------------------------------------------------------------------------------GCT-----CT--AGTCGAGACTGCTCGACTACGAGATCCCCTGAA------------------------------------CCCTTCGCGCAATAGCTTTTTCTATAATTTTTCG |
| droVir3 |
scaffold_12963:12622309-12622520 - |
|
TTTTAATTAAC------A---TTTTTCGTTAAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAAAATTA---------------AACAACTAGCAATTATTTTCTGAGTAATAACCCTGCGAATGACGACTGTATTCTATAAATG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAATACCC---------------------ATTTATTTAAATTTATTTAAAACCATTTAAATAACCATCAACTAAATCCT-----------AAGG-------------CTAATAAGTAAGAG----------GTT-----TT--AGTCGGGAGCTCCCGACTAGGGGATACCCTGAA------------------------------------CCCT------------------------------ |
| droGri2 |
scaffold_15112:347417-347618 - |
|
GTTTTCTTTTA--CAATT-------ATATATACAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------AAATATGATTATATATAGGGAATTGTAAGTCATTGTGTCATTTCAGTTTATTAGCATAC--------------------------------------------------TT------------------------------------------------------AA-------------------------------------------------------------------------------------------TACGCAATTATCAGGGAGAAGTGCTAAACAATGATCTGTTGTAAT-----------TTT-T----------------------------------------------------------------------------------------AAAACAAATAAA-------------------------------------------------------------------AAGTAACAAGACT-------------------------------------------------------------------------------------------------GCT-----TT--AGTCGAGTGAGTTCGACTACGATATTCCCTGGA------------------------------------C--------------------------------- |
| droWil2 |
scf2_1100000004909:11193725-11193935 - |
|
TTTTGTTTTTT--ATG-G-------------------------TTTT---------TG-------------------TTTTATTTGTGATTGTCGAAA------------------------------------------GATA---------------------------------------------------ATAT----------------------------------------------------------------------------AGT-------------AGATATATATACAAATA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TA-TTAAGTCGCCCTATATATATAT-ATTTCACAACATAAGTAAAGAA-----------------------------AAACT---------------------------------------AAC-------------------------------------------ATAAATGAAGTAAGTA--------------------------------------------------------------------------------------------------------------AGTCGTCT--CTCCGACTTCAATATACCATGCACCATGTTTCCATGTTTCCTTTCCGATAGCATTGAAATTCG------------------------------ |
| dp5 |
XR_group8:5628114-5628266 + |
|
AAGATATATACAAAAACAAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAC---AACAAA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAAAAACAAACAAACAAAACAAA-------------------------AAACAAAAGTACTGTACCTTCTTAGCGAA--AAAAACACC---------------------ACACAACTGTACTCAATTCCTGCCCATTAGATGACCTTCATATAGAACCT------------------------------------------------------------------------------------------------------------------------------------------------------------ATAATCCTCTG |
| droPer2 |
scaffold_38:54847-55021 - |
|
TGAAAACCCGT---------------------------------------------------------------------------------------------CG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTATCGAAAGATATATACAAAAACAAAAA-------------------------------------------------------------------------------------------ACAACA-----------------------------------------------------------------------------------------------------------------------AAAAAAAACAAACAAACAAAACAAA-------------------------AAACAAAAGTACTGTACCTTCTTAGCGAAAAAAAAACACC---------------------ACACAACTGTACTCAATTCCTGCCCATTAGATGACCTTCATATAGAACCT------------------------------------------------------------------------------------------------------------------------------------------------------------ATAATCCTCTG |
| droAna3 |
scaffold_10752:2442-2595 + |
|
CATTCATTTTGTATAAAGCA--------------------------------------TACATTTTTCACAA----------------------------------------------TAAATGTATAGCAAAAA----G----------------------AGCGATTAATAAAAGCCAGGCTAACG--ATCT---------------------------------------------------------------------------------GGACAAAATATAAAAA-------------AAAAATATATAACGGAAAAATAAAAATTA-------------------------------------------------------------ATTTTTATACAAA-------------------------------------------------------------A--------------------------------------------AAA-AA------------------------------------TA-AT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAAA------------------------------ |
| droBip1 |
scf7180000395818:45986-46225 + |
|
AAGATTTTTAATTTAAAA------------------ATATTTT-----CTATATGTTG-------------------GC--------------------------------------------------------------------------------------------------------------------------------------------TGCACCAGTTTTTTGTGTTAATTCTTCTTA---ACATGCACCCCCTCACAAAATAA-----------------------------AAAAATTTTTTATAAAAA---------AATAACTAGTTTT------------------------TGGATTAAAAAAATTTCGATAAAACAAT---------------------------------------------------------------------------------------------------------TTTTTGAAAAAAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AACATTTTTAAAGCAAAGAATTTAAGGCACCAAAAAATGG----------GTT-----CTACATCCGATCT-C-TT----TCCAGAACTCCCGCA------------------------------------CTCT------------------------------ |
| droKik1 |
scf7180000302610:901470-901639 + |
|
CAGTAGATATTTGTTGAA---ATATTTGTAAAT----------------------------------------------------------G--------------------------------------AAATG----AAATGAAATAGTTAAATAATTAAAGGAAT-------------------------TGAATA----------------------------------------------------------------------------AAT-------------AAATAAATAAATAAAGA----------------------------------------------------------------------------------------------------------------------------------------------AAAA--------------A--------------------------------------------AAAT-AA-----AAAAATAAATAAATTAAATAAATAAATAGGCAAATA-----------------AATAA-----------------------A-------------------------AAATAA-----------------------------------------AT----------------------------------------------------------------------------------------------------------------------------------------AAATAAATAATTAA---------------------------------------------TAAA------------------------------ |
| droFic1 |
scf7180000453495:172111-172270 - |
|
TATTAAAAACA----GTT-------------------------TCTTTAAAAAAAAAAAAGGATTAGTGTTAAGTTAAATAATACATTATAAGAAATAAAA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTAAAAT-------------AATAAT------------------------------------------------------AA-------------------------------------------------------------------------------------------AATACA---AATAAAAAGAAATGTAAAATAA------------------------------------------------------------------------------------------ATAATAATAATAATAA------------AATACA-------------------------------------------------------------------------AATAAAAAGA-------------------------------------------------------------------------------------------------------------------------------AATGTAAAA---------------------------------------------TTAA------------------------------ |
| droEle1 |
scf7180000490638:1606-1751 - |
|
AGGATATATAAATAAG-T-------------------------TATT---------TA-------------------ATTAATTAATAAATAAATAA-----------------------------------------------------------------ACCAAT-------------------------T-AATA----------------------------------------------------------------------------AAT-------------AAATAAATAGATAAATAAA------------------------------------------------------AA-------------------------------------------------------------------------------------------AAT----------------------------------------------------------AA-----AAAAATATATAAAT-AAATAAATAAATAAGCAAATAA-----------------------------A--------------A-------------------------AAATAA-------------------------------------------ATA-AATAAATAAAAA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGT------------------------------ |
| droRho1 |
scf7180000777251:281835-281965 + |
|
GTTGTCAATCA-------------------------------------------------------CTTTTG-----------------------AAACC---AGAAATCTTTGACAAT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAT------------------------------------------------------AA--------------------------------------------------------------------------------------ACCATATTTTC------------AAACCTTAATTAA------------------------------------------------------------------------------------------ATAACAAAAATAGTAAGA-----------------------------------------------------------------------------------------AACAAGAAAGAAC-------------------------------------------------------------------------------------------------GCT-----AT--AGTCGAGTG-CCTCGACTATTAGATAC--------------------------------------------CC------------------------------ |
| droBia1 |
scf7180000297172:3169-3322 + |
|
TGCAAATGACA--AGGTT-------AGG------------------------------------------------------------GTTATTTAATCCA---AA-----------------------------------AT--------------------------------------------------T-ACTAAATCATGATTATTTAAAAAATATTAAAAATCAAAACTTCTACTTGG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTCAAAGCAGAC--AAAAA-----------------------------------------------------------------------------------------CGAAAAA--------------------------------------------------------------------------------------------GAAACCAAAAAAAGATGAAGACCTGGTGCAGTCT--AATTGAGTA---------------------------------------------------------------------------------------------- |
| droTak1 |
scf7180000413969:47852-48030 + |
|
ATTTTCGTTTC--TGAAT--------------------AACTT-----TTAAA-------------------------------AGTGA-------------------CTTTTTACAGT-AATGTGTAACATATTACAAATTTA---------------------------AAAAGGAAAAGTTAAAGGAATCT---------------------------------------------------------------------------------C------------------------------------------AAGG---------GCTATACAGTTTA------------------------AAGTTTTCAAGAAAATCGTTAGAGCCGT---------------------------------------------------------------------------------------------------------TTTTG-------------------------------------------------------------------AGA--AAAAT-----------------------------------------------------------------------------------------AAAAAAAGCAAAAAAAATTTAAAAAA------------------------------------------------------------------------------------------------------------------------------------C---------------------------------------------CTTT------------------------------ |
| droEug1 |
scf7180000409063:93083-93202 + |
|
ATCAAAAATAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAATTA-------------------------------------------------------------AATTATATGCAAA-------------------------------------------------------------T--------------------------------------------AAA-TT------------------------------------TA-ATAAATATTAAACTGAAT--AAGATCAAAATAAGAACA-----------------------------------------------------------------------------------------AACATGAGGAAAC-------------------------------------------------------------------------------------------------GCT-----AT--AGTCGAGTG-CCGCGACTATAAGATAC--------------------------------------------CC------------------------------ |
| dm3 |
chrX:3447327-3447551 + |
|
TTTATAGAAAT---------------------------------------------TG-------------------AACAACTCATAATTATCAATAAAATTAAA-----------------------------------AT--------------------------------------------------T-TGTA---------------------------------------------------------------------------------------------AACAAACAAATACTAT----------------------------------------------------------------------------------------------------------------------------------------------AAAATATATTGCC---AAT--------------------------------------------ATA----------------------AT---------------TAGTTAAAATATATAATAAAT--AAATA-----------------------------------------------------------------------------------------AGCAAAAATTGGCTCATAGTACAAAA-----T---------------------------------------GTATCATCGCTGAACCGAAGTTGTTAACGAATCGAACAAAGGACATAAATTCATTGATGTGC--AGGCAAGTA-CCTC------------------------------------------------------------------------------ACGAATTTCCG |
| droSim2 |
node_90918:15490-15588 + |
|
AAATTTTATTGTATAAATAA--------------------------------------AACAATCATTTTTG----------------------------------------------AAATAGCATAGC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AA----------------------------------AATAAAA------------------------------------------------------------------AACAAGAGAGAAT-------------------------------------------------------------------------------------------------GCT-----AT--AGTCGAGT--CCTCAACTATTAGA---------------------------------------------CTCC------------------------------ |
| droSec2 |
scaffold_93:39620-39718 - |
|
AAATTTTATTGTATAAATAA--------------------------------------AACAATCATTTTTG----------------------------------------------AAATAGCATAGC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AA----------------------------------AATAAAA------------------------------------------------------------------AACAAGAGAGAAT-------------------------------------------------------------------------------------------------GCT-----AT--AGTCGAGT--CCTCAACTATTAGA---------------------------------------------CTCC------------------------------ |
| droYak3 |
chr2h:218465-218659 + |
|
TAATTTTTTTTTTTTT--------TTAGTATATATAAGT----TTTT----------T-------------------TGTGGTGGGAAATTATTGAAG------------------------------------------TTTG---------------------------------------------------GTGA----------------------------------------------------------------------------GGA-------------AAATATGT--TCATGTGAG----------A------GAATGGTGTTCTGTGTA-------------------------------------------------------------AA-------------------------------------------------------------T--------------------------------------------ATG-TG------------------------------------AA-GTAAATCGAAACAAAAAG--AGGAAAAAAACAGAAAAA-----------------------------------------------------------------------------------------AACAAGAGAGAAC-------------------------------------------------------------------------------------------------GCT-----AT--AGTCGAGTT-CCCCGACTATCTGATAC--------------------------------------------CC------------------------------ |
| droEre2 |
scaffold_4838:201144-201292 - |
|
TTGTTTTTTTA--TTGTT---ACTTTAGCACA----------------------------------------AGTTTATTTATTCTTTATCAAATTCATAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATAAAATAAAT-----AAATAAATAAATTAATA----------------------------------------------------------------------------------------------------------------------------------------------ATAA--------------T--------------------------------------------AAT----------------------TT---------------TAAAAAAAATTTAA-T-----TT----------------------------------AATTAATTTAAAAAAAAATT----------------------------------------AA-------------------TAATAATAATA------------------------------------------------------------------------------------------------------------------------------A---------------------------------------------TAAA------------------------------ |