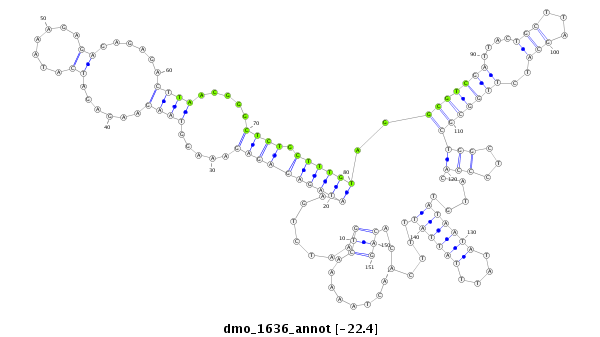
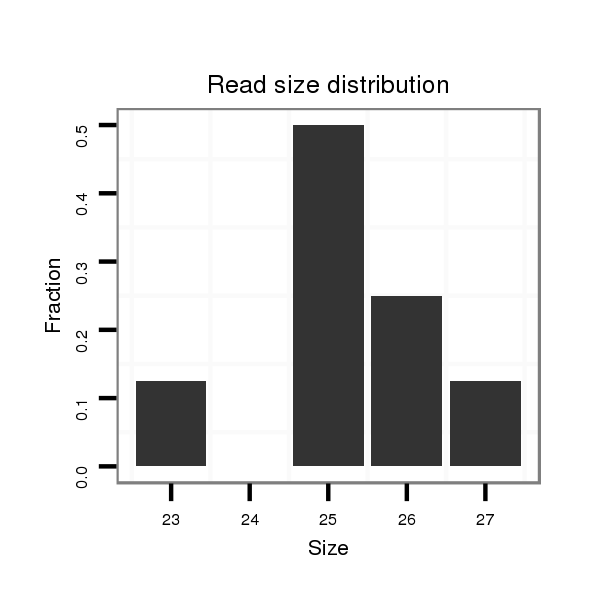
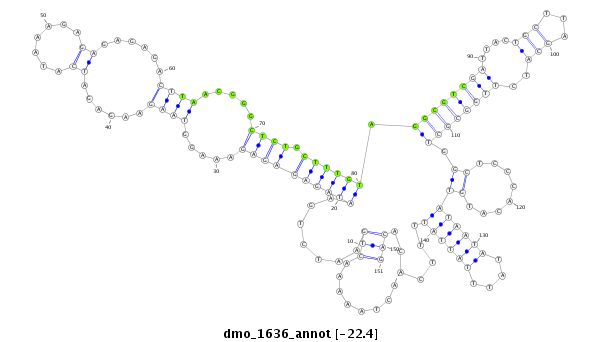
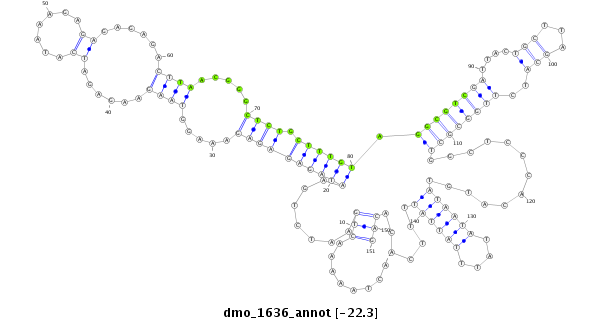
| ACA-----CAACCACAATTTAGATCGTCAACTTATTTATTACACTTCGATGTGATTGCTTCCAAAG---------TCAAAGTCAGACTAATAACTACGAATTAAGTATGTATAACAAAAGAGTACAGTCAAGTAAAGTGTGTGAGAGA-------------------AAGAGAGAGAG---------------------AGCCGTCGATCCTCTCTCA--GCATCGTGGTGCTGGTTCCCACAC---ATATAATGTAT----------------------------------------------------------------------------------- | Size | Hit Count | Total Norm | Total | M026
Head | M043
Female-body | M056
Embryo | V046
Embryo | V052
Head | V058
Head | V120
Male-body | SRR1275488
Male larvae | SRR1275486
Male prepupae | SRR1275484
Male prepupae | GSM1528802
follicle cells |
| ..........................................................................................................................TACAGTCAAGTAAAGTGTGTGAGAG.................................................................................................................................................................................................. | 25 | 9 | 3.56 | 32 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 29 |
| ..........................................................................................................................TACAGTCAAGTAAAGTGTGTGAGAGA-------------------.............................................................................................................................................................................. | 26 | 9 | 2.89 | 26 | 0 | 3 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 18 |
| .................................................TGTGATTGCTTCCAAAG---------TCAAAGTCA................................................................................................................................................................................................................................................................. | 26 | 13 | 1.15 | 15 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| ...............................................................................................................TAACAAAAGAGTACAGTCAAGTAAAG............................................................................................................................................................................................................ | 26 | 9 | 0.89 | 8 | 0 | 5 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| .................................................TGTGATTGCTTCCAAAG---------TCAAAGTC.................................................................................................................................................................................................................................................................. | 25 | 13 | 0.77 | 10 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 |
| ............................................................................................................................................................................................................TCGATCCTCTCTCA--GCATCGTGGTGC............................................................................................................. | 26 | 13 | 0.54 | 7 | 0 | 5 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................TCCTCTCTCA--GCATCGTGGTGCTGGT......................................................................................................... | 26 | 14 | 0.50 | 7 | 0 | 6 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................TACAGTCAAGTAAAGTGTGTGAGA................................................................................................................................................................................................... | 24 | 10 | 0.40 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| ...................TAGATCGTCAACTTATTTATTACACTTCGA.................................................................................................................................................................................................................................................................................................... | 30 | 11 | 0.27 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| ..............................................................................................................................................................................................................GATCCTCTCTCA--GCATCGTGGTGC............................................................................................................. | 24 | 13 | 0.23 | 3 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................TAACAAAAGAGTACAGTCAAGTA............................................................................................................................................................................................................... | 23 | 9 | 0.22 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................TCAACTTATTTATTACACTTCGATG.................................................................................................................................................................................................................................................................................................. | 25 | 11 | 0.18 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................TACACTTCGATGTGATTGCTTCCAAAG---------.......................................................................................................................................................................................................................................................................... | 27 | 12 | 0.17 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................GTCGATCCTCTCTCA--GCATCGTGG................................................................................................................ | 24 | 13 | 0.15 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................TAACAAAAGAGTACAGTCAAGTAAAGT........................................................................................................................................................................................................... | 27 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................ACAGTCAAGTAAAGTGTGTGAGAG.................................................................................................................................................................................................. | 24 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................TAACAAAAGAGTACAGTCAAGTAAAGTG.......................................................................................................................................................................................................... | 28 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................CAGACTAATAACTACGAATTAAGTATG........................................................................................................................................................................................................................................ | 27 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................ACAGTCAAGTAAAGTGTGTGAGAGA-------------------.............................................................................................................................................................................. | 25 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................AACAAAAGAGTACAGTCAAGTAAAGTG.......................................................................................................................................................................................................... | 27 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................AACAAAAGAGTACAGTCAAGTAAAG............................................................................................................................................................................................................ | 25 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................TCAACTTATTTATTACACTTCGATGTGAT.............................................................................................................................................................................................................................................................................................. | 29 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................TATTTATTACACTTCGATGTGAT.............................................................................................................................................................................................................................................................................................. | 23 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................TTATTTATTACACTTCGATGTGAT.............................................................................................................................................................................................................................................................................................. | 24 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................TTAGATCGTCAACTTATTTATTACACT........................................................................................................................................................................................................................................................................................................ | 27 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................TCGATGTGATTGCTTCCAAAG---------TCAAAGT................................................................................................................................................................................................................................................................... | 28 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TGATTGCTTCCAAAG---------TCAAAGTCAGACT............................................................................................................................................................................................................................................................. | 28 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................GCTTCCAAAG---------TCAAAGTCAGACTAA........................................................................................................................................................................................................................................................... | 25 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................TTTAGATCGTCAACTTATTTATTACAC......................................................................................................................................................................................................................................................................................................... | 27 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TCAGACTAATAACTACGAATTAAGT........................................................................................................................................................................................................................................... | 25 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................TTTAGATCGTCAACTTATTTATTACACT........................................................................................................................................................................................................................................................................................................ | 28 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................TTAGATCGTCAACTTATTTATTACAC......................................................................................................................................................................................................................................................................................................... | 26 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................TTCGATGTGATTGCTTCCAAAG---------TCAAAG.................................................................................................................................................................................................................................................................... | 28 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TCAGACTAATAACTACGAATTAAGTA.......................................................................................................................................................................................................................................... | 26 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................TCGTCAACTTATTTATTACACTT....................................................................................................................................................................................................................................................................................................... | 23 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................AGCCGTCGATCCTCTCTCA--GCATCGTGG................................................................................................................ | 28 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................ATGTGATTGCTTCCAAAG---------TCAAAGTC.................................................................................................................................................................................................................................................................. | 26 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................TGTGATTGCTTCCAAAG---------TCAAAG.................................................................................................................................................................................................................................................................... | 23 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................GCCGTCGATCCTCTCTCA--GCATCG................................................................................................................... | 24 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................AGCCGTCGATCCTCTCTCA--GCATCG................................................................................................................... | 25 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................TCGATCCTCTCTCA--GCATCGTGGT............................................................................................................... | 24 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................TGTGATTGCTTCCAAAG---------TCAAAGTCAGA............................................................................................................................................................................................................................................................... | 28 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................TCTCTCA--GCATCGTGGTGCTGG.......................................................................................................... | 22 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................TCCTCTCTCA--GCATCGTGGTGCTGG.......................................................................................................... | 25 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |