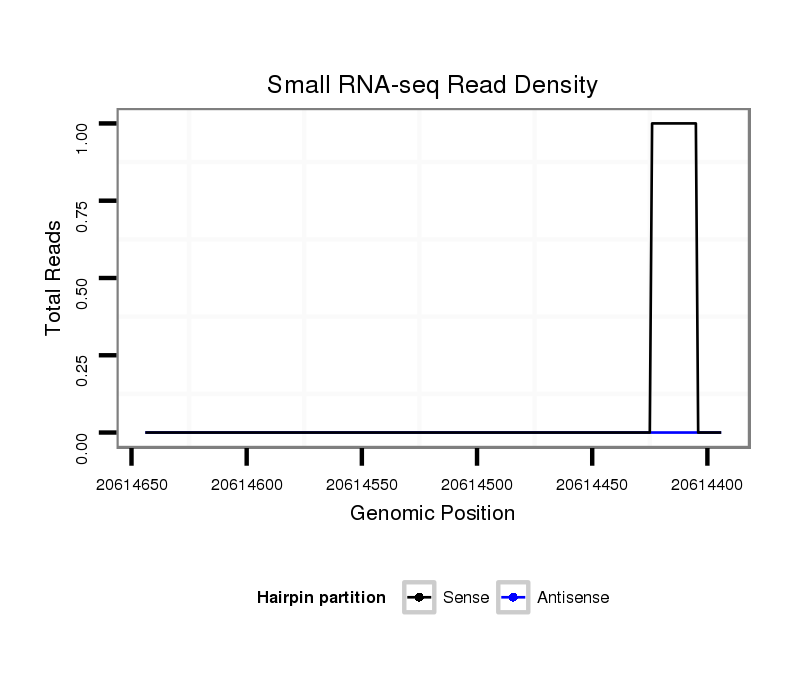

ID:dmo_1609 |
Coordinate:scaffold_6500:20614444-20614594 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dmoj_GLEANR_793:7]; CDS [Dmoj\GI22737-cds]; intron [Dmoj\GI22737-in]
| Name | Class | Family | Strand |
| (TTG)n | Simple_repeat | Simple_repeat | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- AACTAAATCAGCTGGAGAACAATAAGGAGACAGAGAACCTGCAATCATTGGTAAGTAATGCTTTTAATTAGATTGTTATGATGAATGATTCTTGAATCGGTGCTTATCAATGTGATTGTACATGCAGGCCGGCGACAGCGAGCCGCAGACATCTGCCCATGATTCATAGCCGACAACAACAAAGACATCTACAACAACACCCACAACAACAGCAATTGTTGTTCCCCTCGAAGCTGCTGAAGCCTAGCTTA **************************************************....((((((((((..........(((((((...(((............((((..((((...)))).)))).....(((..((....))..)))((((....)))).)))...)))))))..........)))).))).))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V049 head |
V110 male body |
|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................GCCATGATGCATAGCCGAC............................................................................. | 19 | 2 | 1 | 3.00 | 3 | 3 | 0 | 0 |
| ............................................................................................................................................................CCATGATGCATAGCCGAC............................................................................. | 18 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ............................................................................................................................................................................................................................GTTCCCCTCGAAGCTGCTGA........... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...............................................TTGGTAAGTGATGCTTTTAATT...................................................................................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................................................CTCGAAGCTGCTGAACTCT...... | 19 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| ...........................................................................................................................................................GCCATGATGCATAGCCGA.............................................................................. | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 1 |
| ..............................................................................................................................................................................................................CAACATCAATTCTTGTTC........................... | 18 | 2 | 14 | 0.07 | 1 | 1 | 0 | 0 |
| ...................................................................................ACTGAGTCTTGAATCGGG...................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ..............................................................................................................................................................ATGATACGTAGCCGACCA........................................................................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................TAATTGCTGTTCCCCTTGAA................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ............................................................................................................................................................CAATGATACGTAGCCGAC............................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
|
TTGATTTAGTCGACCTCTTGTTATTCCTCTGTCTCTTGGACGTTAGTAACCATTCATTACGAAAATTAATCTAACAATACTACTTACTAAGAACTTAGCCACGAATAGTTACACTAACATGTACGTCCGGCCGCTGTCGCTCGGCGTCTGTAGACGGGTACTAAGTATCGGCTGTTGTTGTTTCTGTAGATGTTGTTGTGGGTGTTGTTGTCGTTAACAACAAGGGGAGCTTCGACGACTTCGGATCGAAT
**************************************************....((((((((((..........(((((((...(((............((((..((((...)))).)))).....(((..((....))..)))((((....)))).)))...)))))))..........)))).))).))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V049 head |
V110 male body |
|---|---|---|---|---|---|---|---|---|---|
| ..........TGACCTCTTGTTATTCCTCT............................................................................................................................................................................................................................. | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................CTTGGACCTTAGTAATCA....................................................................................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| .............................................GTAACCATTCATCAGGAATA.......................................................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................TTGTGGGTTTTGTTATCGTCAA.................................. | 22 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 |
| ............................................................................................................................................................................................................TCGTTGTAGTTAAGAACAAG........................... | 20 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ...................................................................................................................................................................................GTTCCTGTAGATGTTGTTC..................................................... | 19 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6500:20614394-20614644 - | dmo_1609 | AACTAAATCAGCT---GGAGAACAATAAGGAGACAGAGAACCTG----CAATCATTGGTAAGTAATGCTTTTAATTAGATTGTTATG-----AT--------------------------GAATGAT----------------------------------------TCTTGAATCGGTGCTTATCAATGTGATTG------TACATGCAGGCCGGCGACAGCGAGCCGCAGACATCTGCCCATGAT------TCATAGCCGACAACAAC---A------------------------------AAGACATCTACAACAACACCCACAACAACAGCAATT--------GTTGTTCCCCTCGA--AG---------------CTGCTGAAGCCTAGCTTA |
| droVir3 | scaffold_12963:11911285-11911531 - | dvi_11427 | AACTAAACGAGCT---AGAGAATAGTGGGGAGCAGGAGAAACAGCTTTTAACCGTAAGTAATTG----------------------------CATATATATTCAATTTAT-------------CGTA----------------------------------------TTCGA-GAGAGT-----------------TCTTATCAAATTCAGACCAACAAAAGTGAGCCCCAAACACCTGCCCACAGC------ACATAGCTGATAACGAC---A------------------------------ACAACTACAACAACAACAACAACAACAAAAACAACTTGCGACTTGTTAACCACCTCCG--AGC---------AGCCGCCGCTTAAGCATAGTTTA |
| droGri2 | scaffold_15126:5455745-5456060 - | AACTGAAAACGCT---GGAAAGCAATCAGGAGCAACAGCACCAGATGCTTACAGTAAGTAAATG----------------------------TATAAATTAACAATTTATCGTATCTGATGAGTAATTATGTGTGTATTTATATACATATATATATATACCGATGTATACCACAAGAGTTCTTATCAAAATTCTT----TAACAAATGCAGCCCAGCAACAGCGAGTGCAAAGCACCAGCCCCCACA------ACACAGGCGATAACAAA---G------------------------------ACAACCACAACAACAACAATAACA---ACAACAACTTTCGACTAG------AGCCCAAAGAGTCGTTGGTTCCTCCCCTGCTTAAGCATAGTTTA | |
| droAna3 | scaffold_12916:7562689-7562911 - | AGTCCGAGCAGA---CCGAA------AAGGAGTATGAGAGTCTG---GCAGCTGTGAGTATCTGACGGTTTCAATTACAAAATAATC---------------------------------AAACGAT----------------------------------------TCCAATAACTGTC-ATATACATATGTCTGTCATATCAAATTCAGA------AAACCGATACGAAAACAGCTACCCAAACA------AAAGAGTACAAAGCCAA---A------------------------------ACGAAATCAACAACTAGAGGGGCAACTAGACCTA----CAACTAA------ACCTATAAC----------------------------------- | |
| droBip1 | scf7180000396554:179120-179335 - | AGTCGGACCAGAC---GGAA------AAGGAGTTCGAGAGCCAG---GTAGCTGTGAGTATCTGACGGTTTCGATTACAT-------------------------------------------CGAT----------------------------------------TCCAATAAACGT------------CTTTGTTATATGAAATGCAGA------AAACCGATACGAAAACAGCTACCCAAACA------AAAGAGTACAAAGCCAA---A------------------------------ACGAATTCAACAACTAGAGCTGCAACTAGACCAA----CAACACA------ACCTACAAC--A---------AAACCCACAACA----------A | |
| droKik1 | scf7180000302389:327510-327545 - | ACTAGAC-CGACA---GAGATCCAGCGAAGAGCTGAGGAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droEle1 | scf7180000491028:774945-775136 + | AGCAGAGT---G---CCGAG------AAGGAGCATAAACAAATG---GCGTGTGTGAGTATCTGACGCTTTCGATTACCCCGCAATT---------------------------------AAGCGAT----------------------------------------TCCAATAACCCC------------GGTTGTCATATCAAATGCAGA------AAGGCGATACGAAAATACCTACACTAAAA------AGGGAGCCCA---CCAAAGCCACCAAACCTCCTCCTGCAGTCACAGCAACAA------------------------------CA-------------------------------------------------------------- | |
| droRho1 | scf7180000780250:1034-1223 - | GATCCGA------------G------CAGGAGCAAAAACAAATG---GCATGTGTGAGTATCTGACAGTTTCGATTACCGTGCAATT---------------------------------AAGTGAT----------------------------------------TCCAATAACCCC------------GGTTGTCATATCAAATGCAGA------AAAACGATACGAAAACACCTACCCCAAAG------AGGGAGCCCA---CCAAAGCAACCAAACCT---TCCGCAGCCACAGCTAAAA------------------------CAAAATCA-------------------------------------------------------------A | |
| droBia1 | scf7180000302261:1315899-1316093 + | AATCGAG-CAGAGATCCGAG------GAGGAGCGCAAGCAAATG---GCATGTGTGAGTATCTGACGGTTTCGATTACCCCGCAATT---------------------------------AGGCGAT----------------------------------------TCCAATAACACC------------GCTTGTCATACCAAATGCAGA------AAGGCGATGCGAAAACACCTTCCCAAACAAACACAAGGGAGCCCA---GCAAAGCCACCAAAATC---CCCACAGCCACGGCGA------------------------------------------------------------------------------------------------- | |
| droSec2 | scaffold_3:1523264-1523457 - | AGACATTCGA---------G------CAGAAGAAAAATCAAGTG---ACAATTGTGAGTATCTGACGGCTTTGATTGCATCGCAAGTAACCAAT--------------------------AACCGAT----------------------------------------TCCAATAACCCC------------GGTTGTCATATCAAATGCAGG------AATGCGATACGAAAACACCTTCCCAAACA------AGGAAGCCAA---CCAAAGCCACCAAAATC---CCGGGAGCCACGGCAAAAA------------------------------CA-------------------------------------------------------------A |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
Generated: 05/18/2015 at 12:49 AM