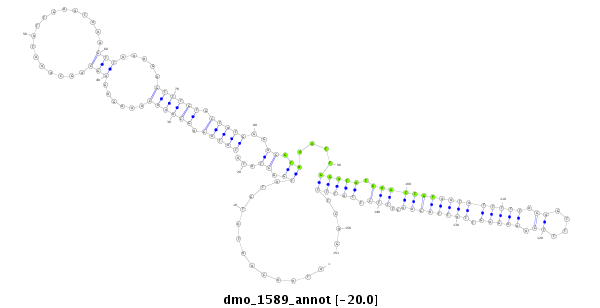

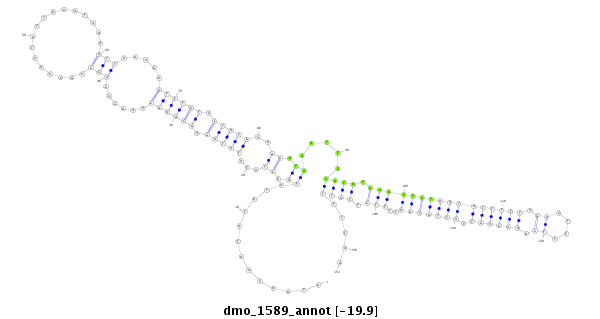
ID:dmo_1589 |
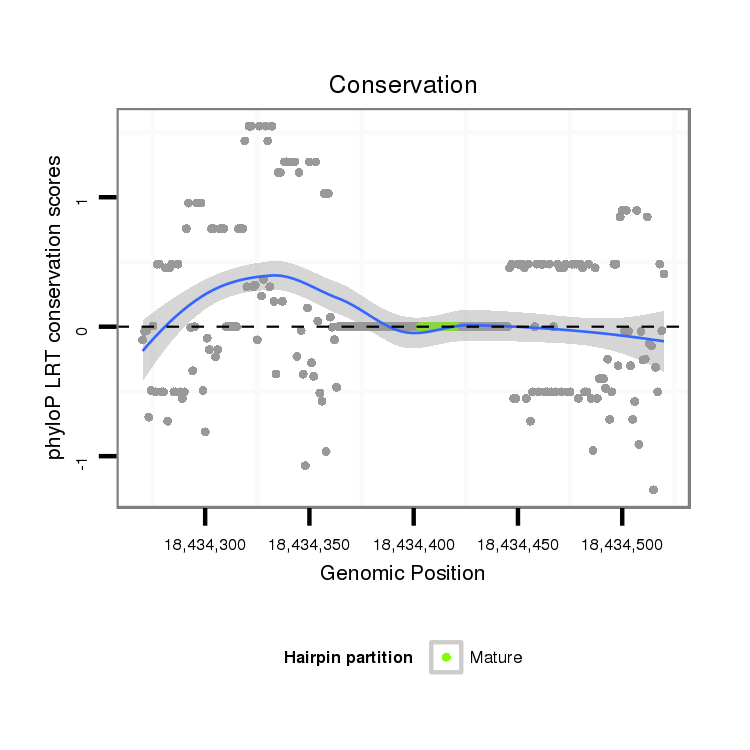
Coordinate:scaffold_6500:18434320-18434470 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -20.0 | -19.9 | -19.9 |
 |
 |
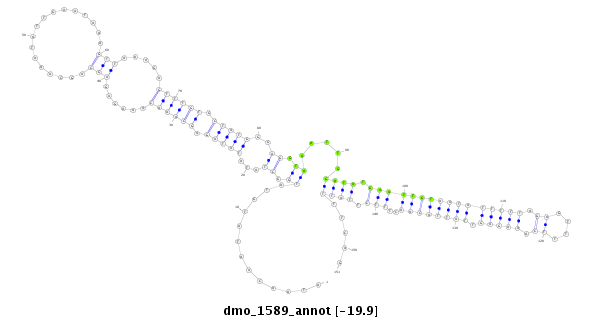 |
CDS [Dmoj\GI17788-cds]; exon [dmoj_GLEANR_2587:3]; intron [Dmoj\GI17788-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ACCAATATAGCCCGAATTCACATTCGTAATGGGTATGAAAAGCATTAAAGATAACAATATGTATGGCTATATATAGAGCAAACAAAGAGAGCAGCAAATATTGAATAAAGTTAAAGAGTTTGTGCTATGGGAGGTAAATTGGATATGAATTGTAATATTTTTAGAGTTTTCAAAGAGTTATTACAACTTTCTTATTTTCAGACTGCATTATGATATGGATTTTGATTTGGAAACGACAACCGCAAATGTCT **************************************************.............((.((...(((((.((((((......(((.................))).....)))))).)))))..)).))....(((((.((((((((((((((((.((....)).))))).))))))))..))).)))))....************************************************** |
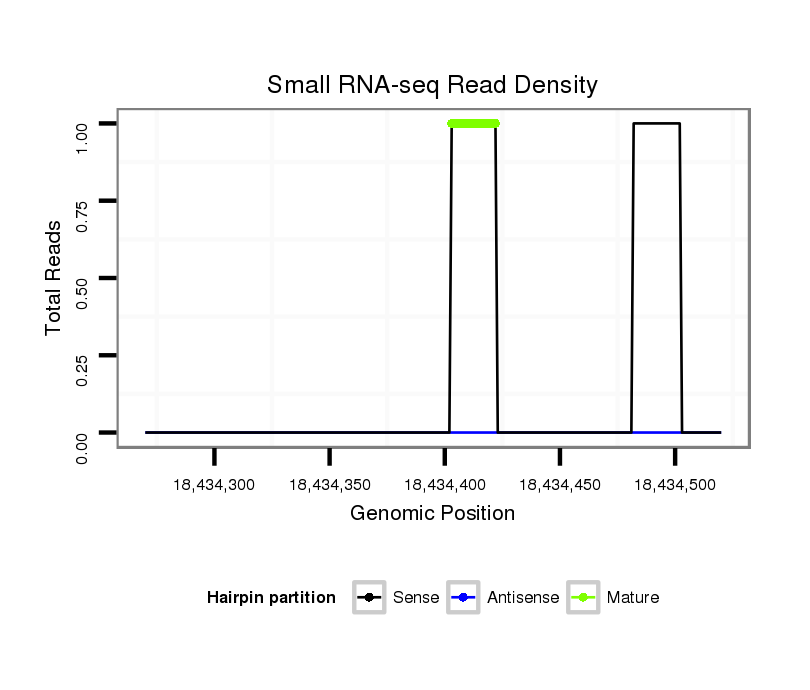
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V041 embryo |
|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................ATATGGATTTTGATTTGGAAA.................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| .....................................................................................................................................GTAAATTGGATATGAATTGT.................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ..............................................................................................................................................................................................................................TGATTTTGAACAGACAACCGC........ | 21 | 3 | 2 | 1.00 | 2 | 2 | 0 |
| ......................................TAAGCATTAAAGATTGCAATATG.............................................................................................................................................................................................. | 23 | 3 | 2 | 0.50 | 1 | 0 | 1 |
| ............CGATTTCACCTTCGTAATGC........................................................................................................................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 1 | 0 |
|
TGGTTATATCGGGCTTAAGTGTAAGCATTACCCATACTTTTCGTAATTTCTATTGTTATACATACCGATATATATCTCGTTTGTTTCTCTCGTCGTTTATAACTTATTTCAATTTCTCAAACACGATACCCTCCATTTAACCTATACTTAACATTATAAAAATCTCAAAAGTTTCTCAATAATGTTGAAAGAATAAAAGTCTGACGTAATACTATACCTAAAACTAAACCTTTGCTGTTGGCGTTTACAGA
**************************************************.............((.((...(((((.((((((......(((.................))).....)))))).)))))..)).))....(((((.((((((((((((((((.((....)).))))).))))))))..))).)))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V110 male body |
V056 head |
|---|---|---|---|---|---|---|---|
| ........TCGAGCTAAAGTGTAAACATTA............................................................................................................................................................................................................................. | 22 | 3 | 7 | 0.29 | 2 | 2 | 0 |
| ..................................TCCTTTTCATAATTTCAATTGT................................................................................................................................................................................................... | 22 | 3 | 5 | 0.20 | 1 | 0 | 1 |
| ........TCGGGCTAAAGTGGAAACAT............................................................................................................................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6500:18434270-18434520 + | dmo_1589 | ACCAATATAGCCCGAATTCACATTCGTAATGGGTATGAAAAGCATTAAAGATAACAATATGTATGGCTATATATAGAGCAAACAAAGAGAGCAGCAAATATTGAATAAAGTTAAAGAGTTTGTGCTATGGGAGGTAAATTGGATATGAATTGTAATATTTTTAGAGTTTTCAAAGAGTTATTACAACTTTCTTATTTTCAGACTGCATTATGATATGGATTTTGATTTGGAAACGACAACCGCAA-ATGTCT |
| droVir3 | scaffold_12963:15476424-15476528 - | TAAGTTGTAATCAGAGCTTTT-TTTGTAAATG------------------------------------------------------------------------------------------------------------------------------------------------GTATTTACTAAC-TTTTGCTC-TAGGCTATATTTTGGCAACGTCCCAACCTTAGAAACACAAGCCACAGATCATAT | |
| droWil2 | scf2_1100000004515:2906827-2906870 + | CAG----------------------------------------------GCTACATATGTGGA------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTGA---AGAGATAACACTGGCCG-AG--GT | |
| droAna3 | scaffold_12903:335435-335501 + | ATATAC---------------ATATATAATATATAAAAAA------AAAGATAAACAAATGTACGGCCATATATGGAATGAGAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------A--AA | |
| droFic1 | scf7180000453982:82760-82811 - | AAA----------------------------------------------GATAAATAAATGTACAGCCATATATGGGGAAAGTAAAAAAAAAGT--------------------------------------------------------------------------------------------------------------------------------------------------------CA--AA | |
| droEle1 | scf7180000490214:1517082-1517133 + | AAA----------------------------------------------GATAAATAAATGTACGGCCATATATAGGGGAAAAAGTAAAAAAAA--------------------------------------------------------------------------------------------------------------------------------------------------------AA--AA | |
| droRho1 | scf7180000779985:465513-465562 - | AAA----------------------------------------------GATAAATAAATGTACAGCCATATATGGGAAAAAAA--GACAAAAA--------------------------------------------------------------------------------------------------------------------------------------------------------CA--AA |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
Generated: 05/18/2015 at 12:46 AM