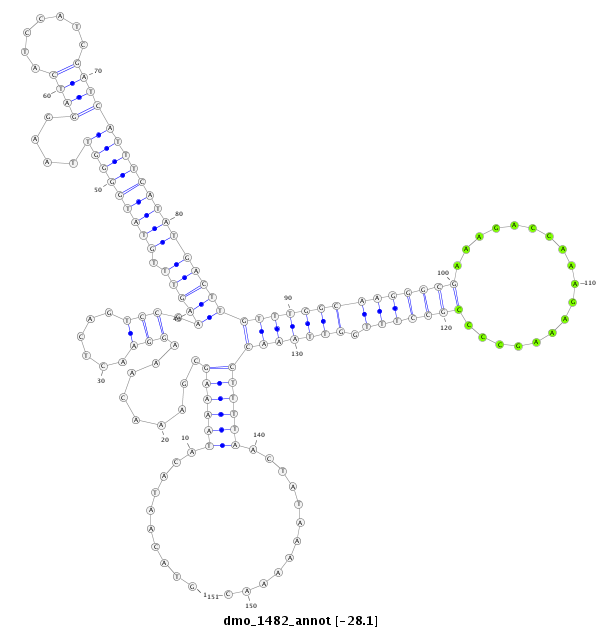
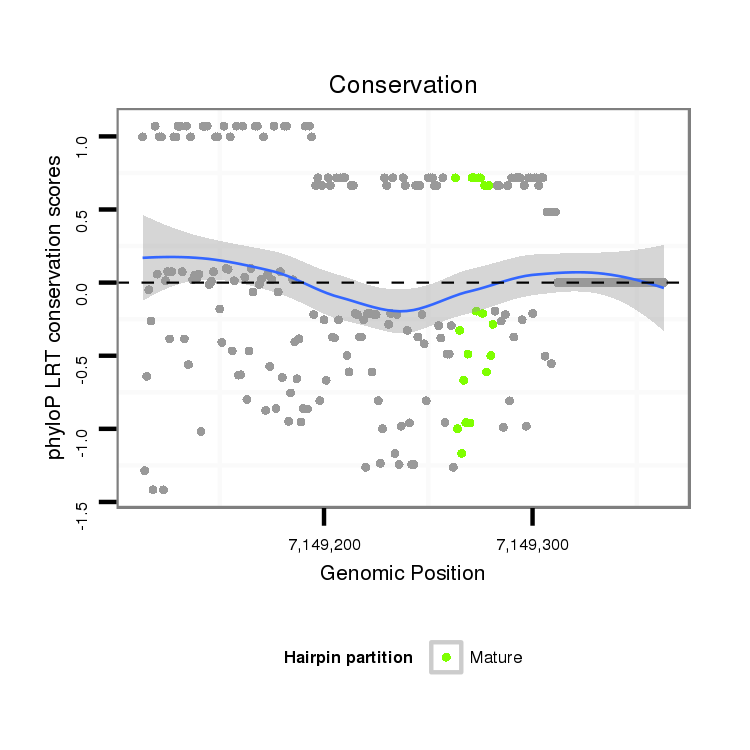
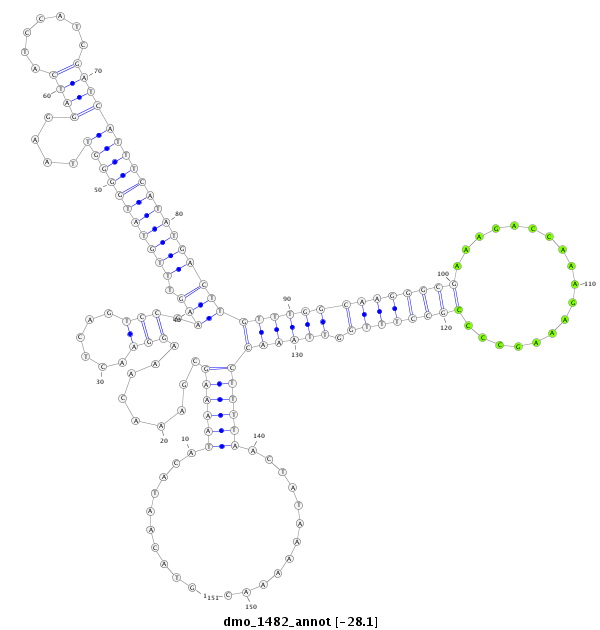
ID:dmo_1482 |
Coordinate:scaffold_6500:7149163-7149313 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -28.1 | -28.1 | -28.1 |
 |
 |
 |
CDS [Dmoj\GI17265-cds]; exon [dmoj_GLEANR_2070:2]; intron [Dmoj\GI17265-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CTACATTAGGACGTGCCAAGATACACAAGTATAAGCGTGAAACTCTCGAGGTACAATACATAAAAGCGAAACAAAGGAACTCAGTCCGAAGTTTGTATGGGGTTAAGGATCATCCATCGATCATTTCATATGACTTGTTTGGCAAGGGCGAAAGACCAAAGAAAGCCCCGCCTTTGGTTAAACCTTTTAACTATAAAAAACTGTTGGAGAGGAATGACGTTGCCAATATGTCACGCCGCAATAAGAGTCAA **************************************************..........((((((.........(((......))).(((((.(((((((((....((((.......))))))))))))))))))((((((((((((((..................))))))).)))))))))))))............************************************************** |
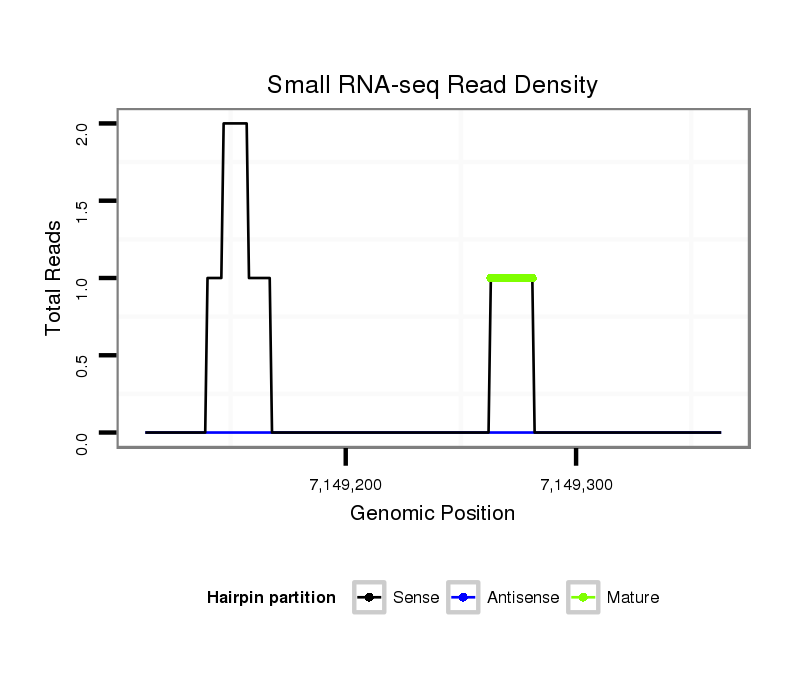
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V110 male body |
M060 embryo |
V056 head |
V049 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ....ATTAAGACGTTCCAAGATACT.................................................................................................................................................................................................................................. | 21 | 3 | 1 | 11.00 | 11 | 2 | 9 | 0 | 0 | 0 |
| .....TTAAGACGTTCCAAGATACT.................................................................................................................................................................................................................................. | 20 | 3 | 3 | 6.33 | 19 | 14 | 4 | 0 | 1 | 0 |
| .....TTAAGACGTTCCAAGATAC................................................................................................................................................................................................................................... | 19 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................AAAGACCAAAGAAAGCCCC.................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................GCGTGAAACTCTCGAGGTACA.................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................ACCAAAGATAACCCCGCCT.............................................................................. | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................AGTATAAGCGTGAAACTC.............................................................................................................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....ATTAAGACGTTCCAAGATA.................................................................................................................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...GATTAAGACGTTCCAAGATA.................................................................................................................................................................................................................................... | 20 | 3 | 5 | 0.40 | 2 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................TTGGCGAGGAAGGACGTCGC............................ | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......AAGACGTTCCAAGATACT.................................................................................................................................................................................................................................. | 18 | 3 | 20 | 0.20 | 4 | 0 | 1 | 0 | 3 | 0 |
| ..................................................................................................................CATCGATCATTTCATATGT...................................................................................................................... | 19 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................GGGTAGGACGTTGCCAATA....................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 |
| ......TAAGACGTTCCAAGATACT.................................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| ..............................................................................................................................................AAAGAGCCAAAGACCAAAGAAA....................................................................................... | 22 | 3 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
|
GATGTAATCCTGCACGGTTCTATGTGTTCATATTCGCACTTTGAGAGCTCCATGTTATGTATTTTCGCTTTGTTTCCTTGAGTCAGGCTTCAAACATACCCCAATTCCTAGTAGGTAGCTAGTAAAGTATACTGAACAAACCGTTCCCGCTTTCTGGTTTCTTTCGGGGCGGAAACCAATTTGGAAAATTGATATTTTTTGACAACCTCTCCTTACTGCAACGGTTATACAGTGCGGCGTTATTCTCAGTT
**************************************************..........((((((.........(((......))).(((((.(((((((((....((((.......))))))))))))))))))((((((((((((((..................))))))).)))))))))))))............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V110 male body |
M060 embryo |
V056 head |
|---|---|---|---|---|---|---|---|---|---|
| ...CTAATTCTGCAAGGTTCTATG................................................................................................................................................................................................................................... | 21 | 3 | 1 | 5.00 | 5 | 1 | 3 | 0 | 1 |
| ....TAATTCTGCAAGGTTCTATG................................................................................................................................................................................................................................... | 20 | 2 | 1 | 5.00 | 5 | 3 | 2 | 0 | 0 |
| ....TAATTCTGCAAGGTTCTAT.................................................................................................................................................................................................................................... | 19 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ....TAATTCTGCAAGGTTCTATGA.................................................................................................................................................................................................................................. | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................CCTCTCCATACTTCAAGGGT.......................... | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................................................CTCTCCATACTTCAAGGGT.......................... | 19 | 3 | 4 | 0.50 | 2 | 0 | 0 | 2 | 0 |
| .....AATTCTGCAAGGTTCTATGA.................................................................................................................................................................................................................................. | 20 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| ...........................TCATAGTCGTACTTGGAGA............................................................................................................................................................................................................. | 19 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................TGGAAAGTTGAAATTCTTTG.................................................. | 20 | 3 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6500:7149113-7149363 + | dmo_1482 | CTACATTAGGACGTGCCAAGATACACAAGTATAAGCGTGAAACTCTCGAGGTACAATACATAAAAGCGAAACAAAGGAACTCAGTCCGAAGTTTGTATGGGGTTAAGGATCATCCATCGATCATTTCATATGACTTGTTTGGCAAGGGCGAAAGACCAAAGAAAGCCCCGCCTTTGGTTAAACCTTTTAACTATAAAAAACTGTTGGAGAGGAATGACGTTGCCAATATGTCACGCCGCAATAAGAGTCAA |
| droVir3 | scaffold_12963:12917033-12917231 - | CAACTGTGGGCCGCGCCAAGGTGCATCGTTATAGGCGTAAAACTCTTGAGATACAATACCAAAACGCAAATAAGCACTATTCGGTGCACAGCTTATATTCGGTTGGACATGACGGCTCAATTGGCTCCACGGGCTACTTTGGCAAAGGCCACCACTATAAGAAAGGCATGCCAATGCTCAAATCCTTTAACTACAATAA---------------------------------------------------- | |
| droGri2 | scaffold_14978:767475-767667 + | CGCGTCTGGGGCGCGCCAAGGTGCGTCGCTATCAGCGCGAAACCCTCAAAGCACAAGTCCACGAAGCAAAGAGACACCAATCGGTCCAAAGTGTATATTCGGCAAAAAGCGTCCATTCGGTGGCATCCGGCGACATGTTTGGACATCCAAATCCAACCAAAAAGGGCACACCACAGGTTAAATCATTCAACTA---------------------------------------------------------- | |
| droWil2 | scf2_1100000004851:614106-614186 + | CCGC-ATGGGTAATACCAAAATTCATCGCTATAAACGCAAGTCAATACAGCAGGAATACGATAATCTCAAACAGAGATATTC------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
Generated: 05/18/2015 at 12:33 AM