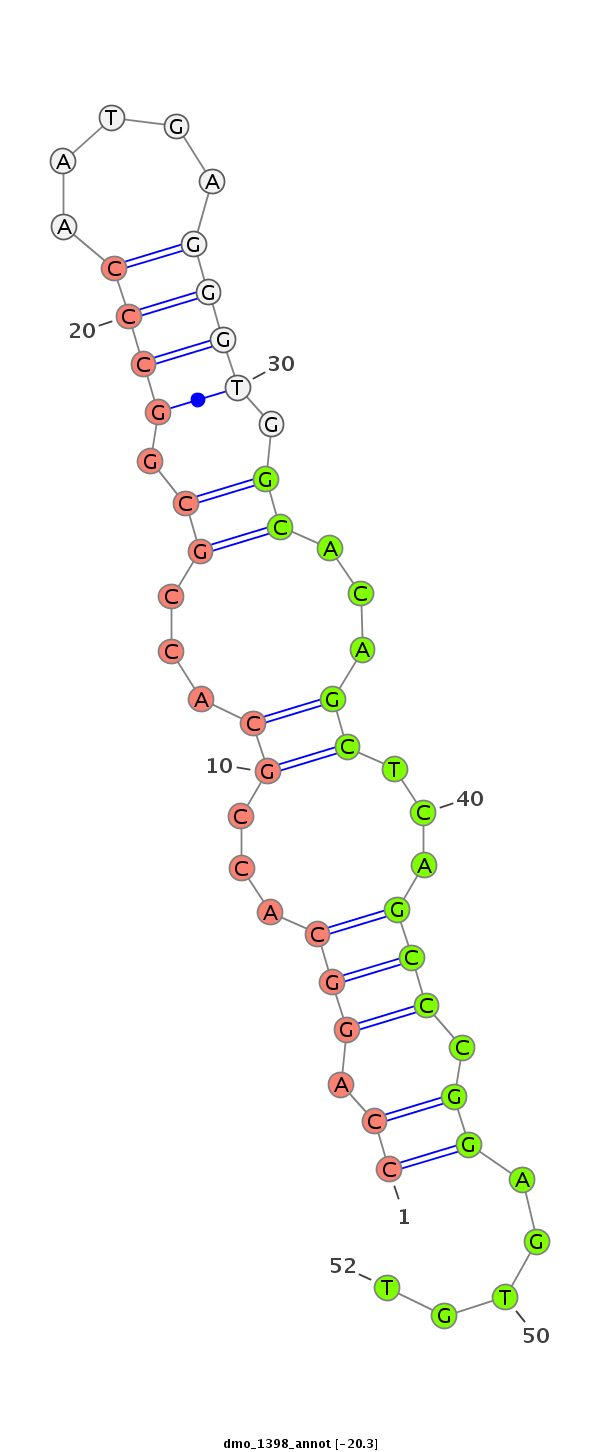
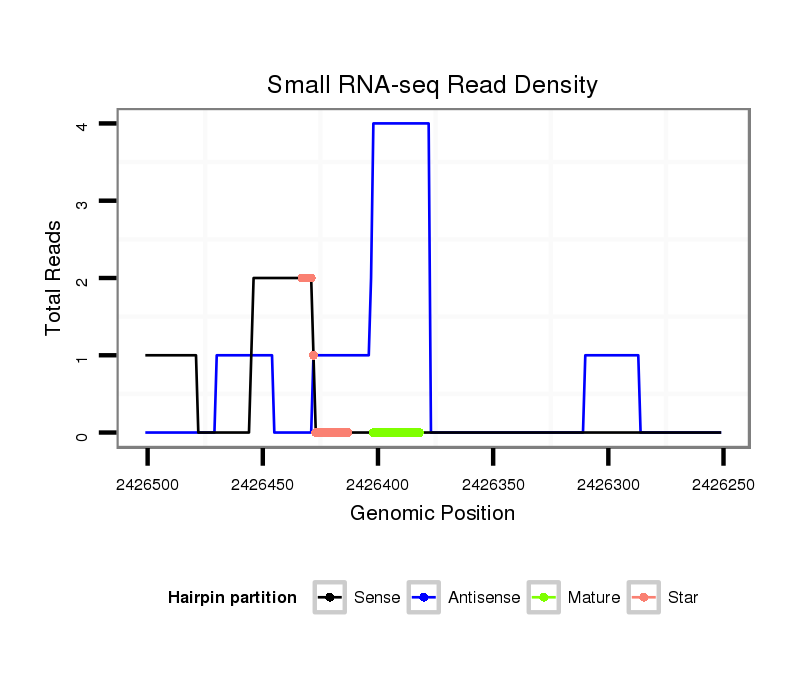
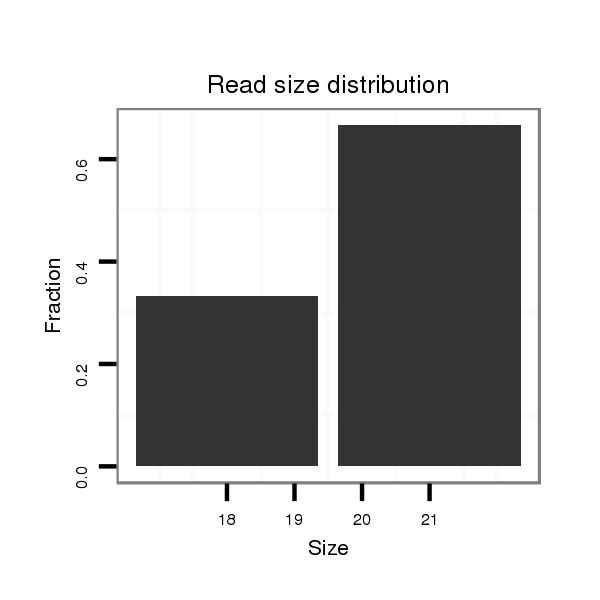

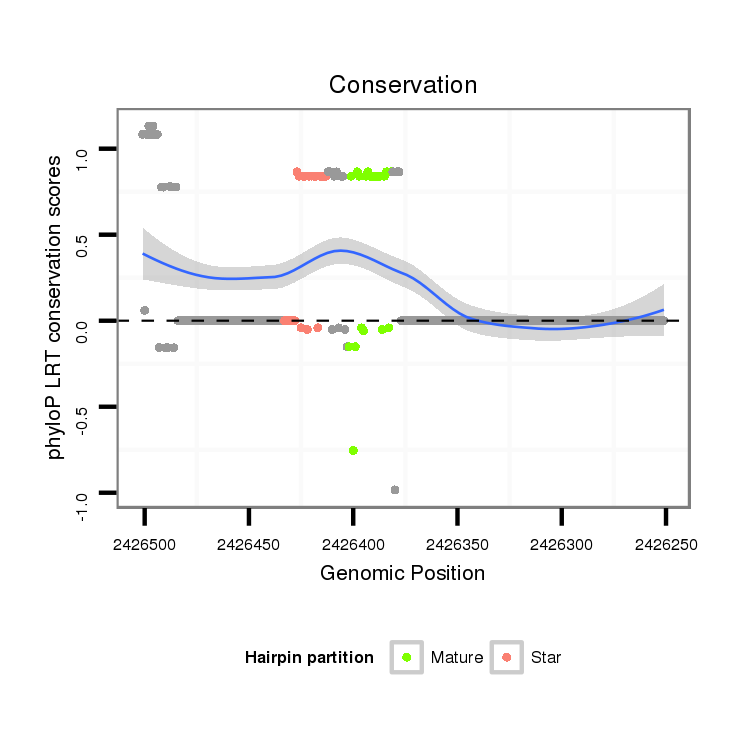
ID:dmo_1398 |
Coordinate:scaffold_6498:2426301-2426451 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
CDS [Dmoj\GI14017-cds]; exon [dmoj_GLEANR_14039:2]; intron [Dmoj\GI14017-in]
No Repeatable elements found
| mature | star |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GGCAGTGGAATCACACTGAAGACTCTGAGTGTCATACGCCAGAGCCCGACGTGAGAAACCACTTGTTTCCAGGCACCGCACCGCGGCCCAATGAGGGTGGCACAGCTCAGCCCGGAGTGTTATACGCCCAGGCTGAGTAGGCAAGGGAACGTGTGCGAGGGGACCAGCAACTGGTCAACTCCGGGAGTCAGGGCGCGGCGGGGACGCATGAAAGATTCCGGGCGTTATACGCCAGAATTTGGACTGGGAAG ********************************************************************((.(((...((...((.((((.....)))).))...))...))).)).....*********************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V110 male body |
V056 head |
V041 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................CGACGTGAGAAACCACTTGTTTCCAGG.................................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................GACGTGAGAAACCACTTGTTTCCAGGC................................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| GGCAGTGGAATCACACTGAAGAC.................................................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................AGGCAAGGGAACGTGAGCC.............................................................................................. | 19 | 2 | 3 | 0.67 | 2 | 0 | 2 | 0 | 0 | 0 |
| ............................................................................................................................................GCAAGGGAACGTGTGCTGG............................................................................................ | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................GCACAGCTCAGCCCGGAGTGT................................................................................................................................... | 21 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................ATGCAAGGGAACGTGTGCTGG............................................................................................ | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................TCAGCCCGGAGTGTTATACGCCCCG........................................................................................................................ | 25 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................................................................ATGCAAGGGAACGTGTGCT.............................................................................................. | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................GGGGGCATAGCTCAGCCGGGAG...................................................................................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................CCAGGCACCGCACCGCGGCCC.................................................................................................................................................................. | 21 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................TCCAGGCACCGCACCGCG...................................................................................................................................................................... | 18 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................TGCCAGGCACCGCACCGCGGC.................................................................................................................................................................... | 21 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................GCGGGTCAACTGCGGGAGT............................................................... | 19 | 3 | 11 | 0.27 | 3 | 0 | 0 | 3 | 0 | 0 |
| ...........................................................................................................................................................................GGGTCAACTGCGGGAGTA.............................................................. | 18 | 3 | 20 | 0.20 | 4 | 0 | 0 | 4 | 0 | 0 |
| ..........................................................................................................................................ATGCAAGGGAACGTGTGCTG............................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| GGCAGTGGAATCAGACTGAAGACC................................................................................................................................................................................................................................... | 24 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................GCCGTGATAAATCACTTGTT........................................................................................................................................................................................ | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................CGGGTCAACGGCGGGAGTC.............................................................. | 19 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................GGTGGCATAGCTCAGCTGGGA....................................................................................................................................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................GACGTGAGAAACCACTGGTTTCC..................................................................................................................................................................................... | 23 | 1 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................AGGCAAGGGAACGTAAGC............................................................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................TTCATATCCGGGAGTCAG............................................................ | 18 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................CGGGTAAACGCCGGGAGT............................................................... | 18 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................GGCATAGCTCAGCTGGGAGTG.................................................................................................................................... | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................GCAAGGGAACGTGAGCC.............................................................................................. | 17 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................GGCACTGCGGTGACGCATGA........................................ | 20 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................TAAGCAAGGGAACGTGAGCT.............................................................................................. | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................................GCAAGGGAACGTGCGCT.............................................................................................. | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................GCAAGGGAACGTGCGCA.............................................................................................. | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................GTAGCAAATGGAACGTGTGC............................................................................................... | 20 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................GGGTCAACGGCGGGAGTC.............................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................CGGGTAAACGCCGGGAG................................................................ | 17 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
|
CCGTCACCTTAGTGTGACTTCTGAGACTCACAGTATGCGGTCTCGGGCTGCACTCTTTGGTGAACAAAGGTCCGTGGCGTGGCGCCGGGTTACTCCCACCGTGTCGAGTCGGGCCTCACAATATGCGGGTCCGACTCATCCGTTCCCTTGCACACGCTCCCCTGGTCGTTGACCAGTTGAGGCCCTCAGTCCCGCGCCGCCCCTGCGTACTTTCTAAGGCCCGCAATATGCGGTCTTAAACCTGACCCTTC
***********************************************************************************************************************************((.(((...((...((.((((.....)))).))...))...))).)).....******************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V041 embryo |
V110 male body |
V056 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................CGTGTCGAGTCGGGCCTCACAATAT............................................................................................................................... | 25 | 0 | 3 | 2.00 | 6 | 4 | 2 | 0 | 0 | 0 |
| ..................................................................................................CCGTGTCGAGTCGGGCCTCACAATAT............................................................................................................................... | 26 | 0 | 3 | 1.67 | 5 | 0 | 3 | 1 | 1 | 0 |
| ...............................................................................................................................................................................................CCGCGCCGCCCCTGCGTACTTTCT.................................... | 24 | 0 | 2 | 1.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...............................................................................................CCACCGTGTCGAGTCGGGCCTCACAATAT............................................................................................................................... | 29 | 0 | 3 | 1.00 | 3 | 1 | 2 | 0 | 0 | 0 |
| .........................................................................GTGGCGTGGCGCCGGGTTACTC............................................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................AGTATGCGGTCTCGGGCTGCACTCT................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................GTCCCGCGCCGCCCCTGCGTACTTTCT.................................... | 27 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................GTATGCGGTCTCGGGCTGCACTCTT.................................................................................................................................................................................................. | 25 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................CCCGCGCCGCCCCTGCGTACTTTCT.................................... | 25 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................TGCGGTCTCGGGCTGCACTCTT.................................................................................................................................................................................................. | 22 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................CAGTCCCGCGCCGCCCCTGCGTACTT....................................... | 26 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................GTCCCGCGCCGCCCCTGCGTACTT....................................... | 24 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................CAGTCCCGCGCCGCCCCTGCGTACT........................................ | 25 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................CGGTCTCGGGCTGCACTCTT.................................................................................................................................................................................................. | 20 | 0 | 6 | 0.50 | 3 | 1 | 1 | 0 | 1 | 0 |
| .............................................................................................................CGGGCCTCACAATATGCGGG.......................................................................................................................... | 20 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................CCACCGTGTCGAGTCGGGCCTCACAA.................................................................................................................................. | 26 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................TGTCGAGTCGGGCCTCACAATAT............................................................................................................................... | 23 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................GTGTCGTCTCGGTCCTCACAAT................................................................................................................................. | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................CCCACCGTGTCGAGTCGGGCCTCACAAT................................................................................................................................. | 28 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................CGGGCCTCACAATATGCGGGT......................................................................................................................... | 21 | 0 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................CCACCGTGTCGAGTCGGGCCTCACAACAT............................................................................................................................... | 29 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................CACCGTGTCGAGTCGGGCCTCACAAT................................................................................................................................. | 26 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................CCCGCGCCGCCCCTGCGTACTT....................................... | 22 | 0 | 6 | 0.33 | 2 | 1 | 1 | 0 | 0 | 0 |
| .....................................CGGTCTCGGGCTGCACTCTTTGGT.............................................................................................................................................................................................. | 24 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................TCCCGCGCCGCCCCTGCGTACTTCCT.................................... | 26 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................CTTATGAGGGTCACAGTAT....................................................................................................................................................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................GTGTCGAGTAGGGGCTCAA.................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6498:2426251-2426501 - | dmo_1398 | GGCAGTGGAATCACACTGAAGACTCTGAGTGTCATACGCCAGAGCCCGACGTGAGAAACCACTTGTTTCCAGGCACCGCACCGCGGCCCAATGAGGGTGGCACAGCTCAGCCCGGA--GTGTTATACGCCCAGGCTGAGTAGGCAAGGGAACGTGTGCGAGGGGACCAGCAACTGGTCAACTCCGGGAGTCAGGGCGCGGCGGGGACGCATGAAAGATTCCGGGCGTTATACGCCAGAATTTGGACTGGGAAG |
| droWil2 | scf2_1100000012052:536-544 + | GGCAGTGGA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_9617:4697-4748 + | --------------------------------------------------------------------------ACTGCGCCGCAGCCCAACGAAGGCTTCGAAGTACAGCCCGGGTAGTATTCTA------------------------------------------------------------------------------------------------------------------------------- | |
| droBip1 | scf7180000395642:5755-5806 + | --------------------------------------------------------------------------ACTGCGCCGCAGCCCAACGAAGGCTTCAAAGTACAGCCCGGGTAGTATTTTA------------------------------------------------------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4770:114231-114247 - | GACAGTGGCATTGCATT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/18/2015 at 12:25 AM