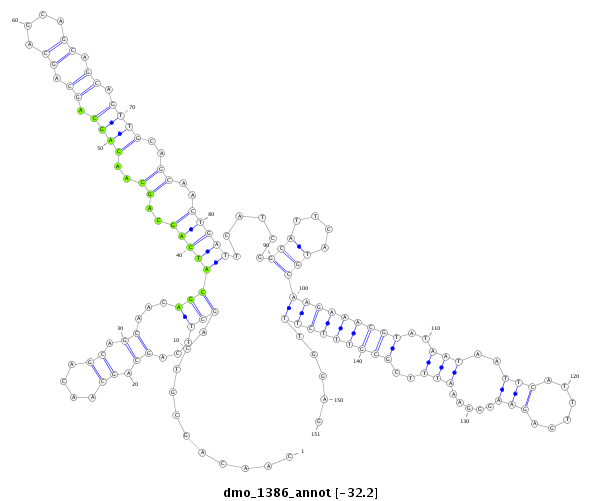
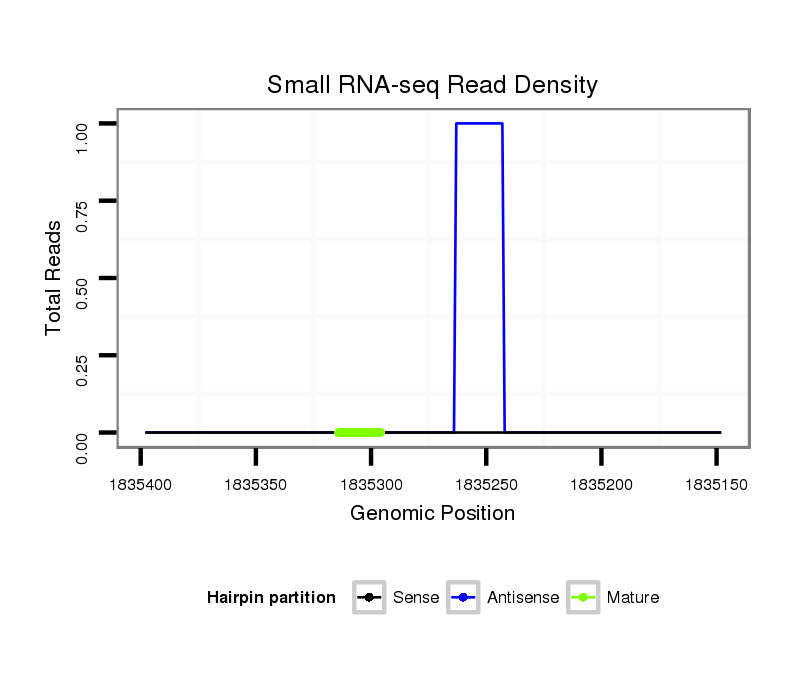
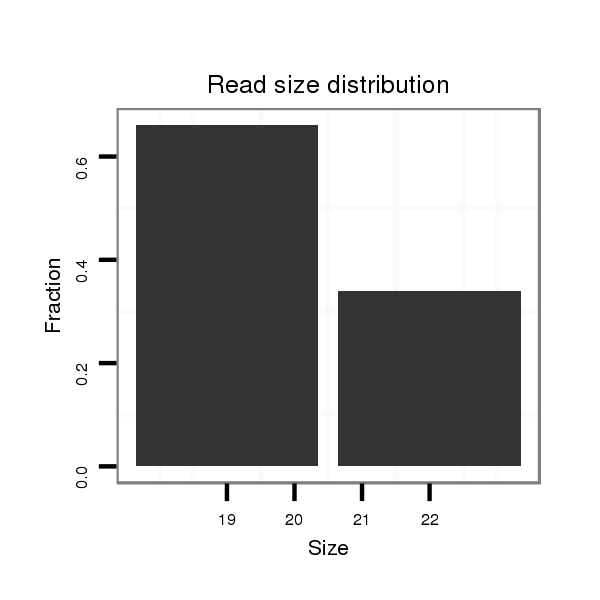
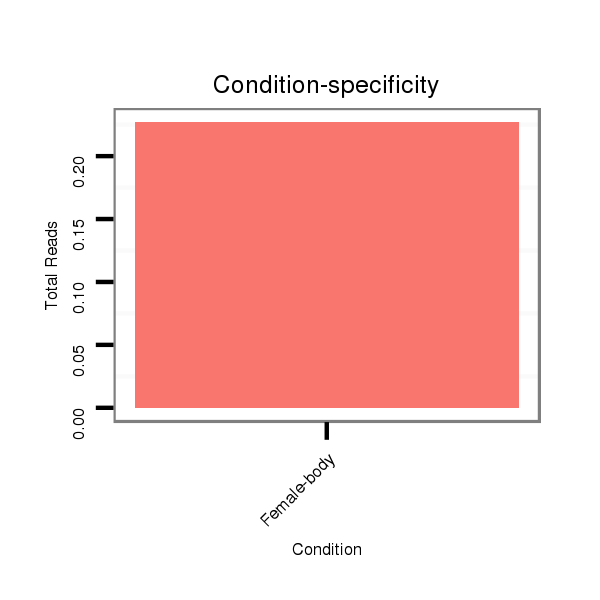
| TGCA---------------GCAGCAT-------------------CAGCAGCAGC---------------AACA--------G----------------------------CA---GCAACAACAGCAGC---AACA------------------------ACAGCAGCAACAACAGCA------------GCAAC------AACAGCAGCAACAACAGCAGCCACAG------CAGCAA---CAGCAGGC--CAC------------------------AGCGGCGGC-------------------------------------------------------------------------AGCAG-----------------------------------------------------------------C--GAGCGTGC-ACAAGTTCATCGACGAACTGACAAAGT------------------------------------------------------------------CCACATCGG------------TGGTCAGCA-----------------------------GCAATGGAACAACAGAGTC-AGC | Size | Hit Count | Total Norm | Total | M022
Male-body | M040
Female-body | M059
Embryo | M062
Head | V043
Embryo | V051
Head | V112
Male-body | SRR902010
Ovary | SRR902011
Testis | SRR902012
CNS imaginal disc |
| .............................................................................................................................................................................................................................................CAA---CAGCAGGC--CAC------------------------AGCGGCGGC-------------------------------------------------------------------------................................................................................................................................................................................................................................................................. | 23 | 1 | 2.00 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................................................GACAAAGT------------------------------------------------------------------CCACATCGG------------TGGTC........................................................ | 22 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................CATCGACGAACTGACAAAGT------------------------------------------------------------------.................................................................................. | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................TCATCGACGAACTGACAAAGT------------------------------------------------------------------CCACA............................................................................. | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................GCAA---CAGCAGGC--CAC------------------------AGCGGC............................................................................................................................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................CAGCAACAACAGCAGCCAC......................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................AGCA------------GCAAC------AACAGCAGCA....................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................AGCAGCAACAACAGCA------------GCA......................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................AGCAGC---AACA------------------------ACAGCAGCA................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................GCA------------GCAAC------AACAGCAGCAAC..................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................G----------------------------CA---GCAACAACAGCAGC---AAC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................GCAGCAACAACAGCA------------GCAAC------................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................GCAGC---AACA------------------------ACAGCAGCAAC............................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................CAACAGCA------------GCAAC------AACAGCAGCAACAAC.................................................................................................................................................................................................................................................................................................................................................................................................................. | 28 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................CAACAGCAGC---AACA------------------------ACAGCAGCAACAAC............................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 28 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................C---------------AACA--------G----------------------------CA---GCAACAACAGCAGC---AACA------------------------AC........................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 28 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CA------------------------ACAGCAGCAACAACAGCA------------GCAAC------AAC.............................................................................................................................................................................................................................................................................................................................................................................................................................. | 28 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................ACA--------G----------------------------CA---GCAACAACAGCAG.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................AC------AACAGCAGCAACAACAGC............................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................CAACAGCAGC---AACA------------------------ACAGCAGCA................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................ACA--------G----------------------------CA---GCAACAACAGCAGC---AAC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................ACAACAGCAGC---AACA------------------------ACAGC..................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................CAGCAGCAACAACAGCA------------GCAAC------A................................................................................................................................................................................................................................................................................................................................................................................................................................ | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................CAACAGCAGC---AACA------------------------ACAGC..................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................ACAGCAGC---AACA------------------------ACAGCAGCAAC............................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................C------AACAGCAGCAACAACAGC............................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................AGCAACAACAGCA------------GCAAC------AACA............................................................................................................................................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................ACAGCAGCAACAACAGCA------------G........................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................ACAGCA------------GCAAC------AACAGCAG......................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................CAACAGCA------------GCAAC------AACAGC........................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CA------------------------ACAGCAGCAACAACAGCA------------GCA......................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................AGCAGCAGC---------------AACA--------G----------------------------CA---GCAA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................ACAGCAGCAACAACAGCAGC............................................................................................................................................................................................................................................................................................................................................................................................................ | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................A------------GCAAC------AACAGCAGCAACAACA................................................................................................................................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................CA------------GCAAC------AACAGCAGCAACAA................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................GCAGCAGC---------------AACA--------G----------------------------CA---GCAAC.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................ACAGCAGCAACAACAGCA------------GCAAC------................................................................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................AGCAGC---------------AACA--------G----------------------------CA---GCAACA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CA------------------------ACAGCAGCAACAACAGC......................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................ACAGCA------------GCAAC------AACAGCAGCAAC..................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................ACAGCAGC---AACA------------------------ACAGCAGC.................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................ACAGCA------------GCAAC------AACAGCAGC........................................................................................................................................................................................................................................................................................................................................................................................................................ | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................CAGCAACAACAGCA------------GCAAC------AA............................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................CAGCA------------GCAAC------AACAGCAGCAACA.................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................ACAGCAGCAACAACAGCA------------GC.......................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................ACA--------G----------------------------CA---GCAACAACAGCAGC---...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................ACAACAGCA------------GCAAC------AACAGC........................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................CA---GCAACAACAGCAGC---AACA------------------------A......................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................CAACAGCA------------GCAAC------AACAGCAGCA....................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................CA--------G----------------------------CA---GCAACAACAGCAGC---AACA------------------------.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................ACAGCAGC---AACA------------------------ACAGCAG................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................A---GCAACAACAGCAGC---AACA------------------------ACA....................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................ACAGCAGCAACAACAGCAG............................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................AGCAGCAGC---------------AACA--------G----------------------------CA---GCA.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................AGC---AACA------------------------ACAGCAGCAACAACA........................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................C---------------AACA--------G----------------------------CA---GCAACAACAGCAGC---A..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................ACA------------------------ACAGCAGCAACAACAGC......................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................CAGC---AACA------------------------ACAGCAGCAACAA............................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |