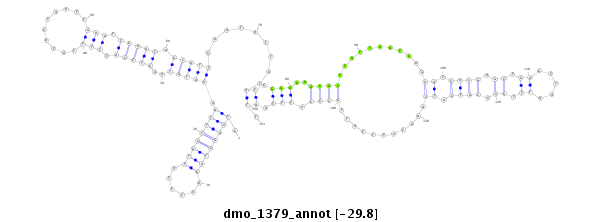
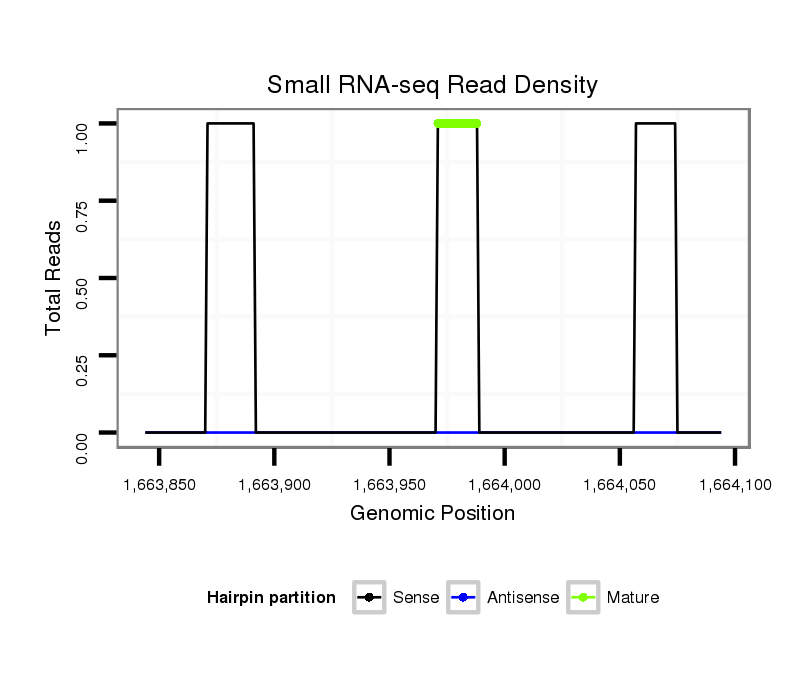
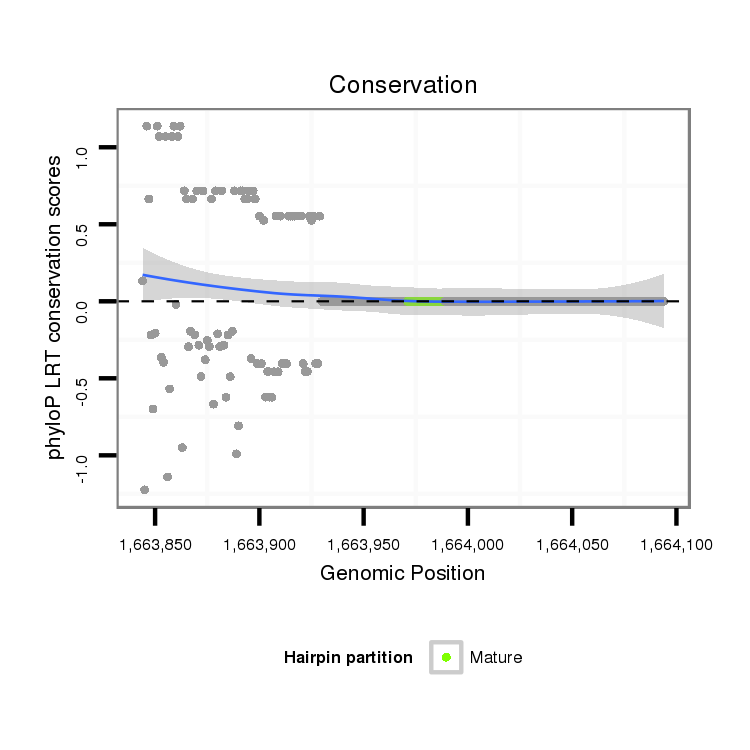
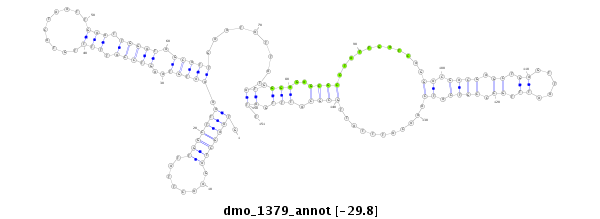
ID:dmo_1379 |
Coordinate:scaffold_6498:1663894-1664044 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -29.8 | -29.8 | -29.8 |
 |
 |
 |
exon [dmoj_GLEANR_14198:3]; CDS [Dmoj\GI14119-cds]; intron [Dmoj\GI14119-in]
| Name | Class | Family | Strand |
| Homo6 | DNA | hAT-Pegasus | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TTAGTCCAGCAGTAGATGAATCCGCAACCAAGCCAATCACTTCGATGAAGGTAAGCTAGAACTTATTGGCTTAAAATGTAAGTCCATTTTTGTACTAATTGAATTGGATAGCATTGAATATTATATGGGAGAGCGCCTATTGATCAGGACGACGAGCTGAGTTAATTTGCCCGTCATCAAACATTTATTGCGCATTTGATTTATTCTGCCAAGGCAGTTAAAGGAAATACGTACAGATGTTGGCTCTTTCT **************************************************.((((((((......)))))))).(((((..(((((.(((..........))).))))).))))).........((.(((..((((...........((.((((.((.((.....)).)).)))).))...........)))).))).)).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V110 male body |
M046 female body |
M060 embryo |
|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................GCCAATCCAGTTAAAGGAAAGA...................... | 22 | 3 | 1 | 29.00 | 29 | 27 | 2 | 0 |
| ...............................................................................................................................................................................................................GCCAATCCAGTTAAAGGAAAG....................... | 21 | 3 | 2 | 21.00 | 42 | 40 | 2 | 0 |
| ..............................................................................................................................................................................................................AGCCAATCCAGTTAAAGGAAA........................ | 21 | 3 | 5 | 4.60 | 23 | 4 | 19 | 0 |
| ...............................................................................................................................................................................................................GCCAATCCAGTTAAAGGAAA........................ | 20 | 2 | 1 | 3.00 | 3 | 0 | 3 | 0 |
| ...............................................................................................................................................................................................................GCCAATCCAGTTAAAGGA.......................... | 18 | 2 | 4 | 2.25 | 9 | 0 | 9 | 0 |
| ...............................................................................................................................................................................................................GCCAATCCAGTTAAAGGAA......................... | 19 | 2 | 2 | 1.00 | 2 | 1 | 1 | 0 |
| .....................................................................................................................................................................................................................GCAGTTAAAGGAAATACG.................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 |
| ...............................................................................................................................GGAGAGCGCCTATTGATC.......................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 |
| ...........................CCAAGCCAATCACTTCGATGA........................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ...........................................GTTGATGGTAAGCTGGAACT............................................................................................................................................................................................ | 20 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................CAATCCAGTTAAAGGAAA........................ | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 1 |
| ................................................................................................................................................................................................................CCAATCCAGTTAAAGGAAAGA...................... | 21 | 3 | 12 | 0.08 | 1 | 0 | 1 | 0 |
| .......................................................................................................................CTCATATGGGAGAGCACCT................................................................................................................. | 19 | 3 | 18 | 0.06 | 1 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................CAATCCAGTTAAAGGAAAGA...................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
|
AATCAGGTCGTCATCTACTTAGGCGTTGGTTCGGTTAGTGAAGCTACTTCCATTCGATCTTGAATAACCGAATTTTACATTCAGGTAAAAACATGATTAACTTAACCTATCGTAACTTATAATATACCCTCTCGCGGATAACTAGTCCTGCTGCTCGACTCAATTAAACGGGCAGTAGTTTGTAAATAACGCGTAAACTAAATAAGACGGTTCCGTCAATTTCCTTTATGCATGTCTACAACCGAGAAAGA
**************************************************.((((((((......)))))))).(((((..(((((.(((..........))).))))).))))).........((.(((..((((...........((.((((.((.((.....)).)).)))).))...........)))).))).)).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V110 male body |
|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................CGGTTAGGTCAATTTCCTTTC....................... | 21 | 3 | 2 | 5.50 | 11 | 11 | 0 | 0 |
| ...............................................................................................................................................................................................................CGGTTAGGTCAATTTCCTT......................... | 19 | 2 | 2 | 3.00 | 6 | 6 | 0 | 0 |
| ................................................................................................................................................................................................................GGTTAGGTCAATTTCCTTT........................ | 19 | 2 | 1 | 3.00 | 3 | 3 | 0 | 0 |
| ..............................................................................................................................................................................................................TCGGTTAGGTCAATTTCCTTT........................ | 21 | 3 | 5 | 2.80 | 14 | 14 | 0 | 0 |
| ..........................................................................................................................................................TCGACTCAATTAAACGGTCAGT........................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................CGGTTAGGTCAATTTCCTTTCT...................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ................................................................................................................................................................................................................GGTTAGGTCAATTTCCTTTC....................... | 20 | 3 | 17 | 0.65 | 11 | 11 | 0 | 0 |
| ................ACTTGGGCGTTGGTTCGG......................................................................................................................................................................................................................... | 18 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ...........CATCTACTTGGGTGTTGG.............................................................................................................................................................................................................................. | 18 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................................TAAGACGGTTCCGGCAATGG............................. | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................GTTAGGTCAATTTCCTTTCT...................... | 20 | 3 | 20 | 0.25 | 5 | 5 | 0 | 0 |
| ................................................................................................................................................................................................................GGTTAGGTCAATTTCCTTTCT...................... | 21 | 3 | 12 | 0.25 | 3 | 3 | 0 | 0 |
| ...............................................................................................................................................................................................................CGGTTAGGTCAATTTCCT.......................... | 18 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| ........................................................................................................................................................................................................ACTAAGACGGTTCGGGCAA................................ | 19 | 3 | 20 | 0.15 | 3 | 0 | 1 | 2 |
| ...............................................................................................................................................................TCAATTAAACAGGCAGTT.......................................................................... | 18 | 2 | 9 | 0.11 | 1 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................GTTAGGTCAATTTCCTTT........................ | 18 | 2 | 12 | 0.08 | 1 | 1 | 0 | 0 |
| .................CTTAGGCGTTAGTTCAGTTG...................................................................................................................................................................................................................... | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 1 |
| ......................GCGTTTGTTGGGGTAGTGA.................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ...................TGGGCGTTGGTTCGGCAA...................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6498:1663844-1664094 + | dmo_1379 | TTAGTCCAGCAGTAGATGAATCCGCAACCAAGCCAATCACT--------------------TC-GATGAAGGTAAGCTAGAACTTATTGGCTTAAAATGTAAGTCCATTTTTGTACTAATTGAATTGGATAGCATTGAATATTATATGGGAGAGCGCCTATTGATCAGGACGACGAGCTGAGTTAATTTGCCCGTCATCAAACATTTATTGCGCATTTGATTTATTCTGCCAAGGCAGTTAAAGGAAATACGTACAGATGTTGGCTCTTTCT |
| droVir3 | scaffold_13052:1321648-1321705 + | TAAGCACAGTTGCAGATGAGTCCGCGATCAAACCCATCAT-----------------TAGCTC-GAACAAGGTGAG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_14822:342737-342843 + | TCAGCATAGCTGGCGAGGATTCAACGACGACAACAACAACGTTCAATAACAAACCATTGGGAGGAACGAAGGTAAGTTGGCTAGAAATAATTTAAAATAATAGTTTA--------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droBia1 | scf7180000302087:340464-340481 + | AGA--CTAGTAGCGGATGAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
Generated: 05/18/2015 at 12:19 AM