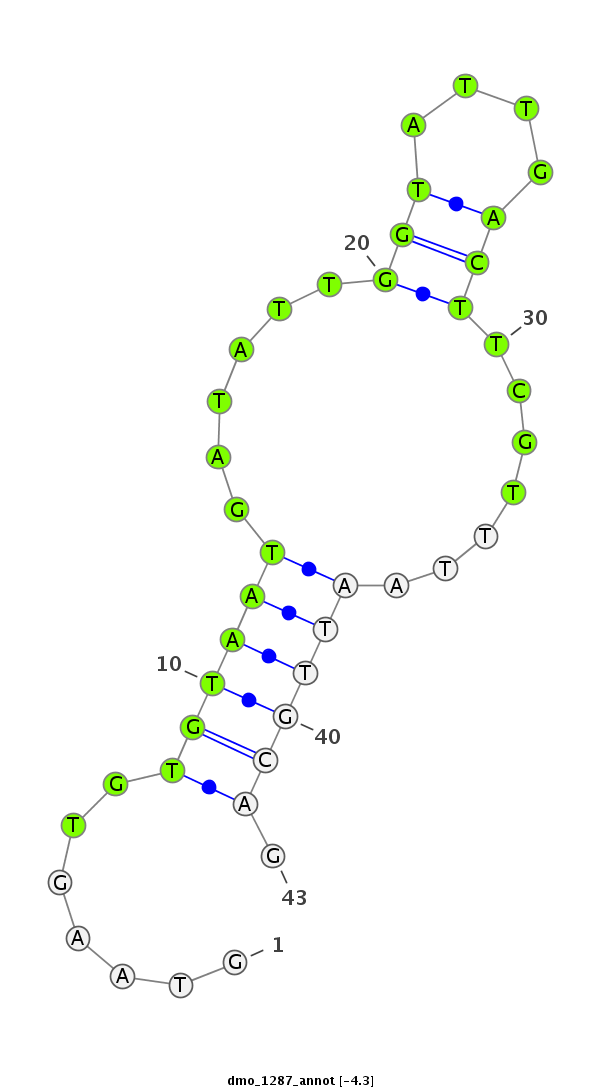
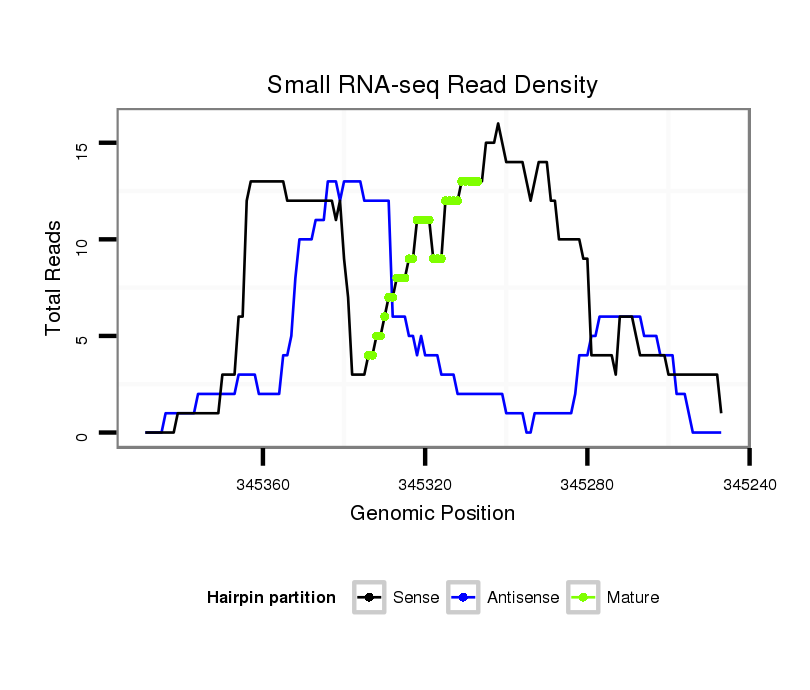
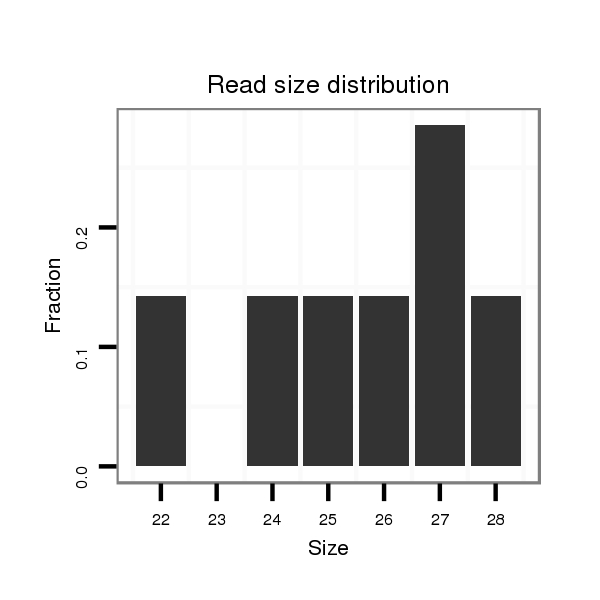

ID:dmo_1287 |
Coordinate:scaffold_6498:345297-345339 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -4.1 | -3.9 | -3.5 |
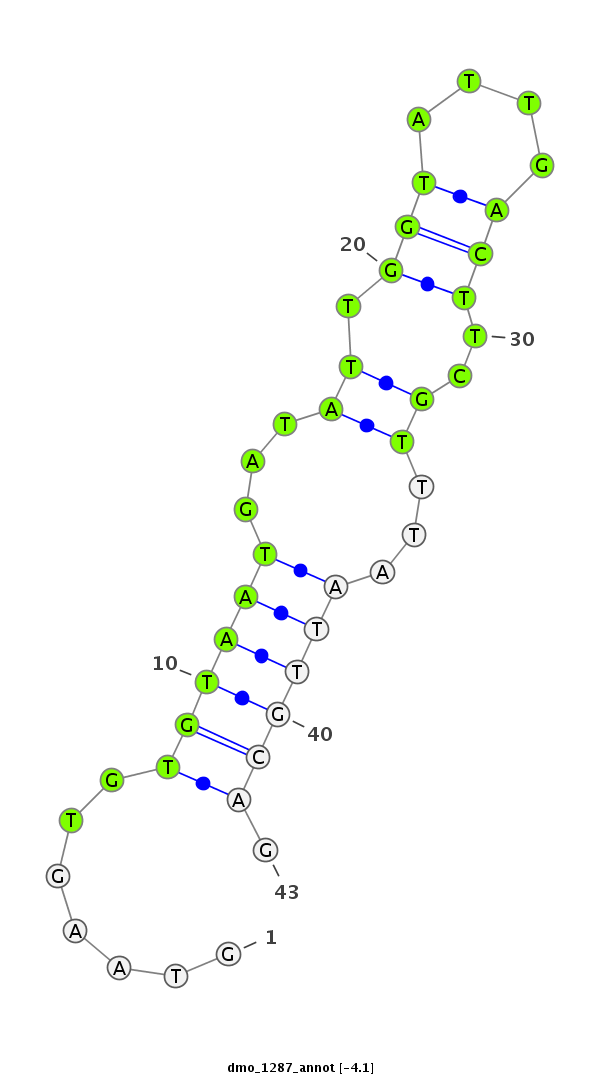 |
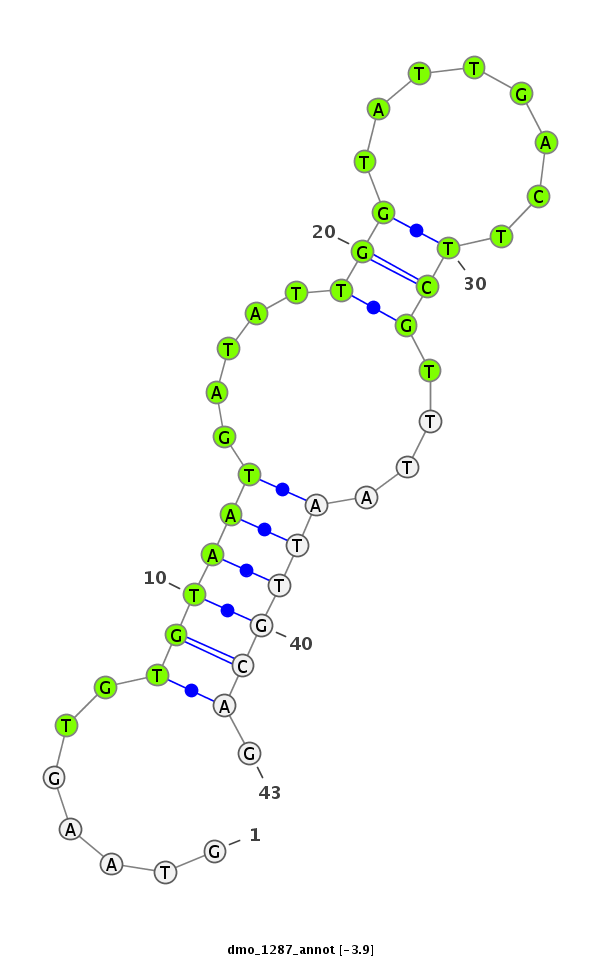 |
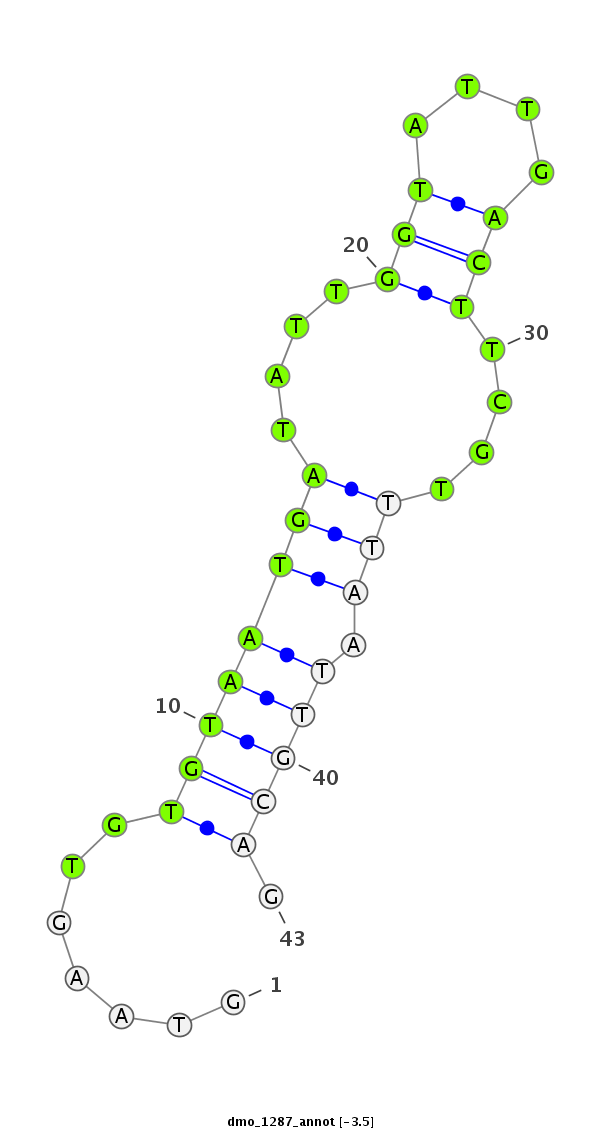 |
intron [Dmoj\GI14068-in]; CDS [Dmoj\GI14068-cds]; exon [dmoj_GLEANR_14116:6]; intron [Dmoj\GI14068-in]; CDS [Dmoj\GI14068-cds]; exon [dmoj_GLEANR_14116:5]
No Repeatable elements found
| ##################################################-------------------------------------------###########################----------------------- GATTCAGCAAATCAACTGCTCCATCTTGGTCATCCAGGGACGGGACATAGGTAAGTGTGTAATGATATTGGTATTGACTTCGTTTAATTGCAGGTATTGGACTTCTGGCATCGGACATAGGTAAGTGTGTAATAAAATTGGAC **************************************************.......((((((......(((....))).......)))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V110 male body |
|---|---|---|---|---|---|---|---|---|
| .........................TTGGTCATCCAGGGACGGGACATAGG............................................................................................ | 26 | 0 | 1 | 4.00 | 4 | 3 | 1 | 0 |
| ....................................................................................TAATTGCAGGTATTGGACTTCTGGCA................................. | 26 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 |
| .....................................................................................................................TAGGTAAGTGTGTAATAAAATTGGA. | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| .......................TCTTGGTCATCCAGGGACGGGACATA.............................................................................................. | 26 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ...................TCCATCTTGGTCATCCAGGGACGGGACA................................................................................................ | 28 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ..........................................................................TGACTTCGTTTAATTGCAGGTATTGG........................................... | 26 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 |
| ..............................................................................TTCGTTTAATTGCAGGTATTGGACTTC...................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ................................................................ATATTGGTATTGACTTAGGT........................................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 |
| ..........................................................................TGACTTCGTTTAATTGCAGGTATTGGAC......................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...............................................TAGGTAAGTGTGTAATGATATTGG........................................................................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ................................................................................................TTGGACTTCTGGCATCGGACATAGGT..................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .......................................................TGTGTAATGATATTGGTATTGACTTCGT............................................................ | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...................................................................TTGGTATTGACTTCGTTTAATTGCAGG................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ................................................AGGTAAGTGTGTAATGATATTGG........................................................................ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .......................................................................................TTGCAGGTATTGGACTTCTGGCATCGGAC........................... | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .........................TTGGTCATCCAGGGACGGGACATAG............................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ...................................................................................TTAATTGCAGGTATTGGACTTCTGG................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...................................................................TTGGTATTGACTTCGTTTAATTGCAGGT................................................ | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................................................................................TGGACTTCTGGCATCGGACATAGG...................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .......................TCTTGGTCATCCAGGGACGGGACATAG............................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ............................................................AATGATATTGGTATTGACTTCG............................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..........................TGGTCATCCAGGGACGGGACATAGG............................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...........................................................TAATGATATTGGTATTGACTTCGTT........................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| .........................TTGGTCATCCAGGGACGGGACATA.............................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ....................................................................................TAATTGCAGGTATTGGACTTCTGGA.................................. | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................................................TATTGGTATTGACTTCGTTTAATT...................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .....................................................................................................................TAGGTAAGTGTGTAATAAAATTGGAC | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ........AAATCAACTGCTCCATCTTGGTCATCC............................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .........................TTGGTCATCCAGGGACGGGACATAGGC........................................................................................... | 27 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ..............................................................TGATATTGGTATTGACTTCGTTTAAT....................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .........................................................TGTAATGATATTGGTATTGACTTCGTT........................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..................................................................................TTTAATTGCAGGTATTGGACTTCTGGCA................................. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...................................................TAAGTGTGTAATGATATTGGTATTGAC................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .........................................................................................................TGGCATCGGACATAGGTAAGTGTG.............. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..............................................................................TTCGTTTAATTGCAGGTATTGGAC......................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
|
CTAAGTCGTTTAGTTGACGAGGTAGAACCAGTAGGTCCCTGCCCTGTATCCATTCACACATTACTATAACCATAACTGAAGCAAATTAACGTCCATAACCTGAAGACCGTAGCCTGTATCCATTCACACATTATTTTAACCTG
**************************************************.......((((((......(((....))).......)))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V041 embryo |
|---|---|---|---|---|---|---|---|---|
| .....................................CCTGCCCTGTATCCATTCACACAT.................................................................................. | 24 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 |
| ....................................CCCTGCCCTGTATCCATTCACACAT.................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ......................................CTGCCCTGTATCCATTCACACAT.................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ................................................................................................................CCTGTATCCATTCACACATTATT........ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..................................GTCCCTGCCCTGTATCCATTCACACAT.................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..........................................CCTGTATCCATTCACACATTACTAT............................................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ..............................................................................................................AGCCTGTATCCATTCACACATTAT......... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..................................GTCCCTGCCCTGTATCCATT......................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..........................................................................................................CCGTAGCCTGTATCCATTCACACAT............ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ....................................................................ACCATAACTGAAGCAAATTAACGTCC................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................................................ATAACCATAACTGAAGCAAATTAA...................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .............................................GTATCCATTCACACATTACTATAA.......................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..............CGACGAGGTAGAACCAGTAGGTCCCT....................................................................................................... | 26 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| .............................................GTATCCATTCACACATTACTATAACCAT...................................................................... | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .........................................CCCTGTATCCATTCACACATTACG.............................................................................. | 24 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| .................................................CCATTCACACATTACTATAACCATAACT.................................................................. | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ........................................GCCCTGTATCCATTCACACATTACT.............................................................................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ......................................CTGCCCTGTATCCATTCACACATTACT.............................................................................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...........................................................................................................CGTAGCCTGTATCCATTCACACAT............ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...........................................................................................................CGTAGCCTGTATCCATTCAC................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .......................AGAACCAGTAGGTCCCTGCCCTGTA............................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .....TCGTTTAGTTGACGAGGTAGAAC................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 |
| ................................................................................................AACCTGAAGACCGTAGCCTGTATCCAT.................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .............TTGACGAGGTAGAACCAGTAGGTCCCT....................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6498:345247-345389 - | dmo_1287 | GATTCAGCAAATCAACTGCTCCATCTTGGTCATCCAGGGACGGGACATAGGTAAGTGTGTAATGATATTGGTATTGACTTCGTTTAATTGCAGGTATTGGACTTCTGGCATCGGACATAGGTAAGTGTGTAATAAAATTGGAC |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
Generated: 05/18/2015 at 12:04 AM