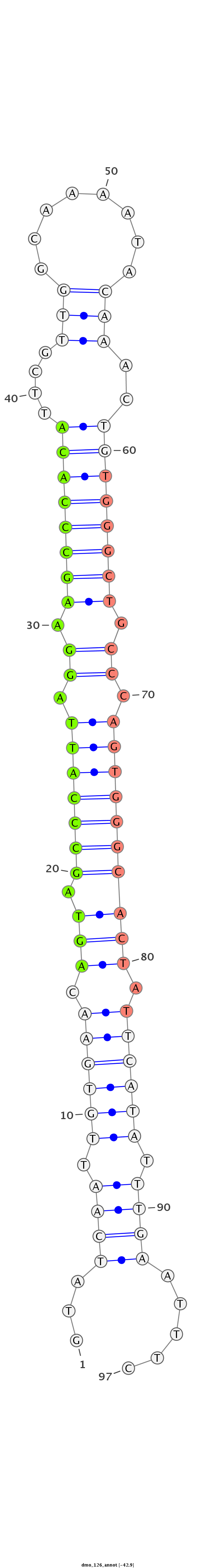

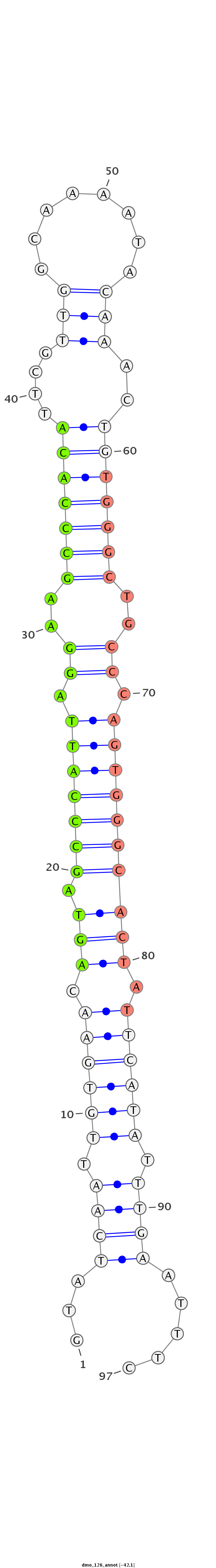
ID:dmo_126 |
Coordinate:scaffold_6328:2302261-2302329 - |
Confidence:confident |
Class:Canonical miRNA |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
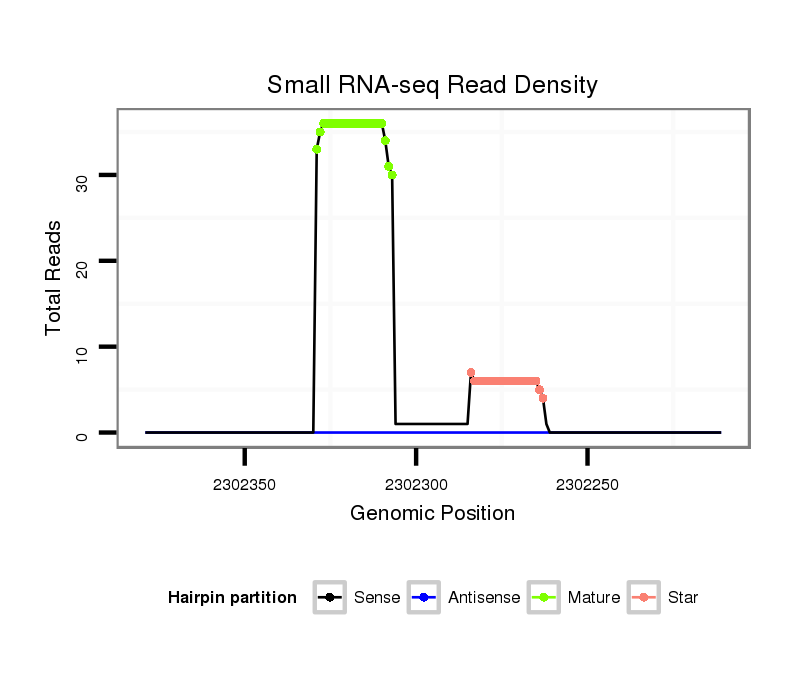
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -42.1 |
 |
intergenic
No Repeatable elements found
|
CGCAGGTTTATAAAAGCCAATCAGGTGAACTATGTGTATCAATTGTGAACAGTAGCCCATTAGGAAGCCCACATTCGTTGGCAAAATACAAACTGTGGGCTGCCCAGTGGGCACTATTCATATTTGAATTTCCCGCCAACCAATTGACAATTAACTTGGACTTGATAAT
***********************************...((((.((((((.(((.(((((((.((.((((((((....(((........)))..)))))))).)).)))))))))).)))))).)))).....************************************* |
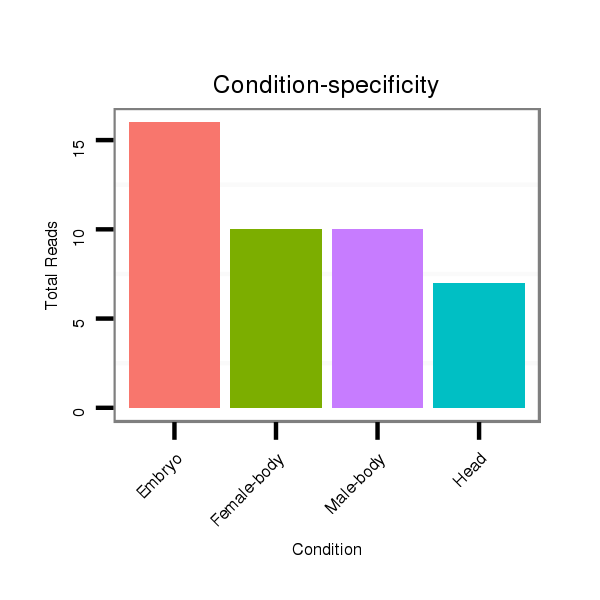
Read size | # Mismatch | Hit Count | Total Norm | Total | M060 embryo |
V110 male body |
M046 female body |
V056 head |
V049 head |
V041 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................AGTAGCCCATTAGGAAGCCCACA................................................................................................ | 23 | 0 | 1 | 26.00 | 26 | 11 | 7 | 3 | 1 | 3 | 1 |
| ..................................................AGTAGCCCATTGGGAAGCCCACA................................................................................................ | 23 | 1 | 1 | 5.00 | 5 | 0 | 2 | 0 | 3 | 0 | 0 |
| ...............................................................................................TGGGCTGCCCAGTGGGCACTAT.................................................... | 22 | 0 | 1 | 3.00 | 3 | 1 | 0 | 1 | 1 | 0 | 0 |
| ..................................................AGTAGCCCATTAGGAAGCCCA.................................................................................................. | 21 | 0 | 1 | 3.00 | 3 | 0 | 2 | 0 | 0 | 1 | 0 |
| ..................................................ATTAGCCCATTAGGAAGCCCACA................................................................................................ | 23 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGTAGCCCATTAGGAAGCCC................................................................................................... | 20 | 0 | 1 | 2.00 | 2 | 0 | 0 | 1 | 1 | 0 | 0 |
| ..................................................AGTAGCCCATTAGGAAGCCCAC................................................................................................. | 22 | 0 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 |
| ...................................................GTAGCCCATTAGGAAGCCCACA................................................................................................ | 22 | 0 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................AGTAGCCCATTAGAAAGCCCACA................................................................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................AGCAGCCCATTAGGAAACCCACA................................................................................................ | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................TGGGCTGCCCAGTGGGCACTATAA.................................................. | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................ATTCGTTGGCAAAATACAAACTGT......................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................TGGGCTGCCCAGTGGGCACT...................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................AGTAGCCCATTGGGAAGCCCA.................................................................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ATTAGCCCATTAGGAAGCCCAC................................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................TGGGCTGCCCAGTGGGCACTA..................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................TGGGCTGCCCAGTGGGCACTATT................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................AGTAGCCCATTAGGAAGCCCATC................................................................................................ | 23 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................AGTAGCCCATTGGGAAGCCCAC................................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................TTGGCTGCCCAGTGGGCACTAT.................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................TAGCCCATTAGGAAGCCCACA................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AATAGGCCATTAGGATGCCCA.................................................................................................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
|
GCGTCCAAATATTTTCGGTTAGTCCACTTGATACACATAGTTAACACTTGTCATCGGGTAATCCTTCGGGTGTAAGCAACCGTTTTATGTTTGACACCCGACGGGTCACCCGTGATAAGTATAAACTTAAAGGGCGGTTGGTTAACTGTTAATTGAACCTGAACTATTA
*************************************...((((.((((((.(((.(((((((.((.((((((((....(((........)))..)))))))).)).)))))))))).)))))).)))).....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droMoj3 | scaffold_6328:2302211-2302379 - | dmo_126 | confident | CGCAGGTTTATAAAAGCCAATCAGGTGAACTATGTGTATCAATTGTGAACAGTAGCCCATTAGGAAGCCCACATTCGTTGGCAAAATACAAACTGTGGGCTGCCCAGTGGGCACTATTCATATTTGAATTTCCCGCCAACCAATTGACAATTAACTTGGACTTGATAAT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
Generated: 10/20/2015 at 07:28 PM