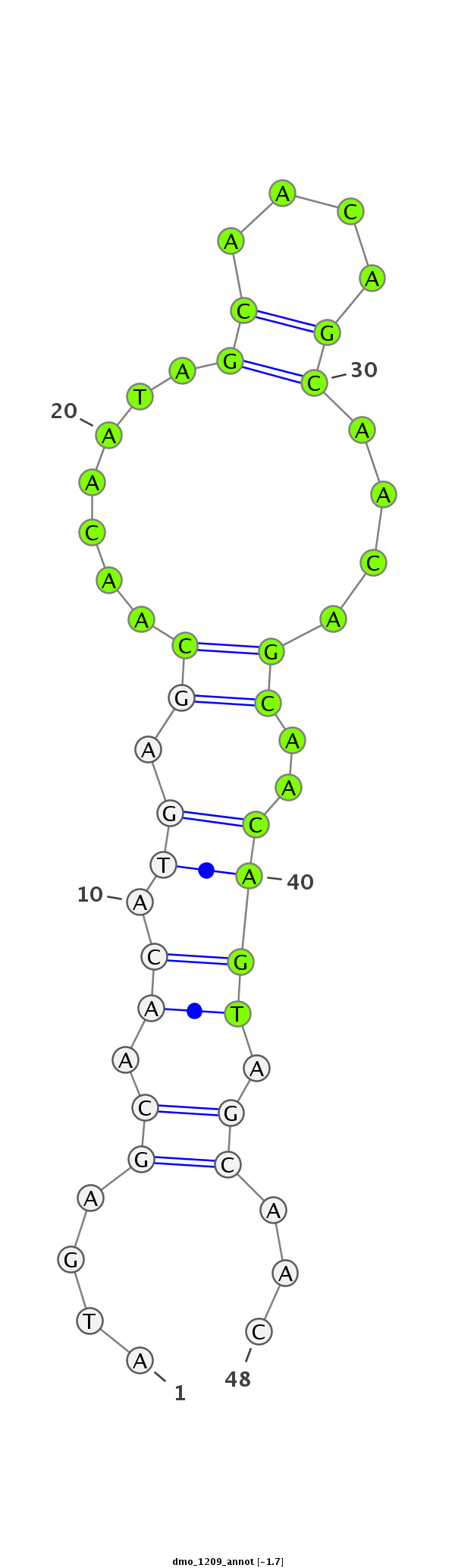
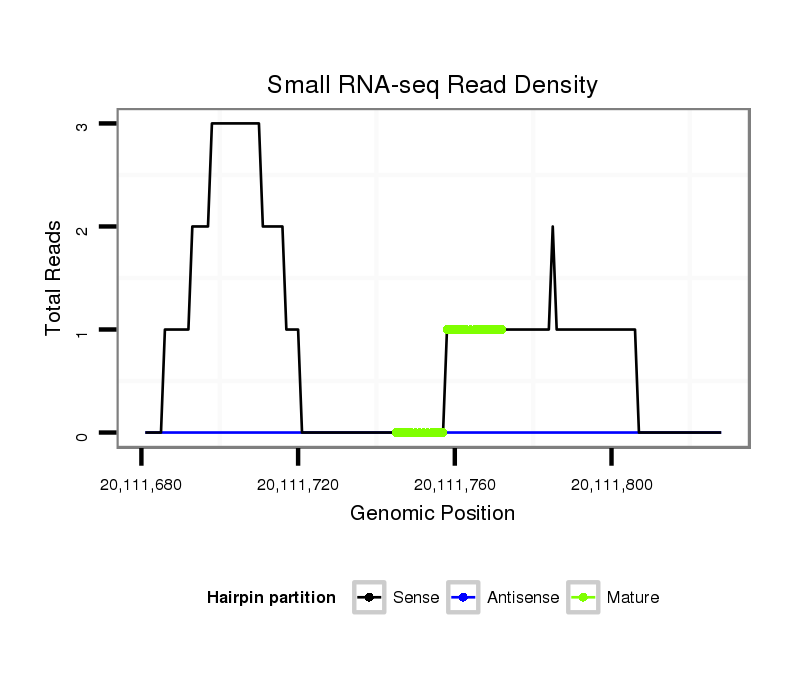
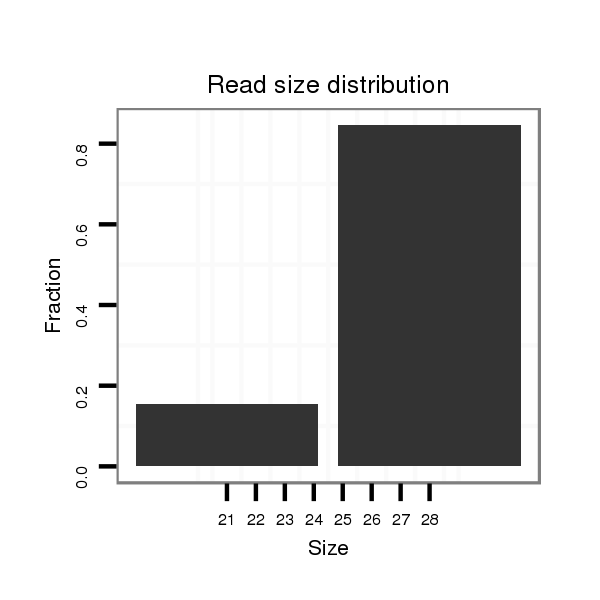
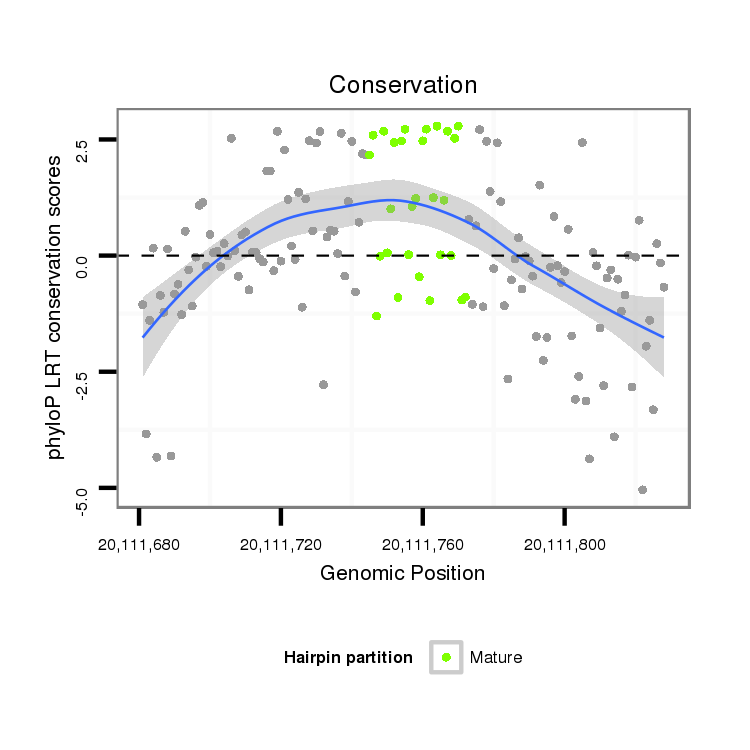
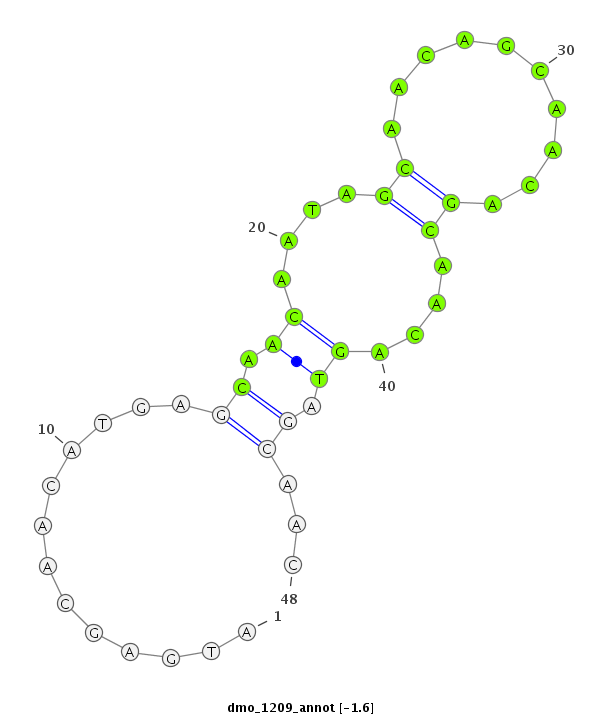
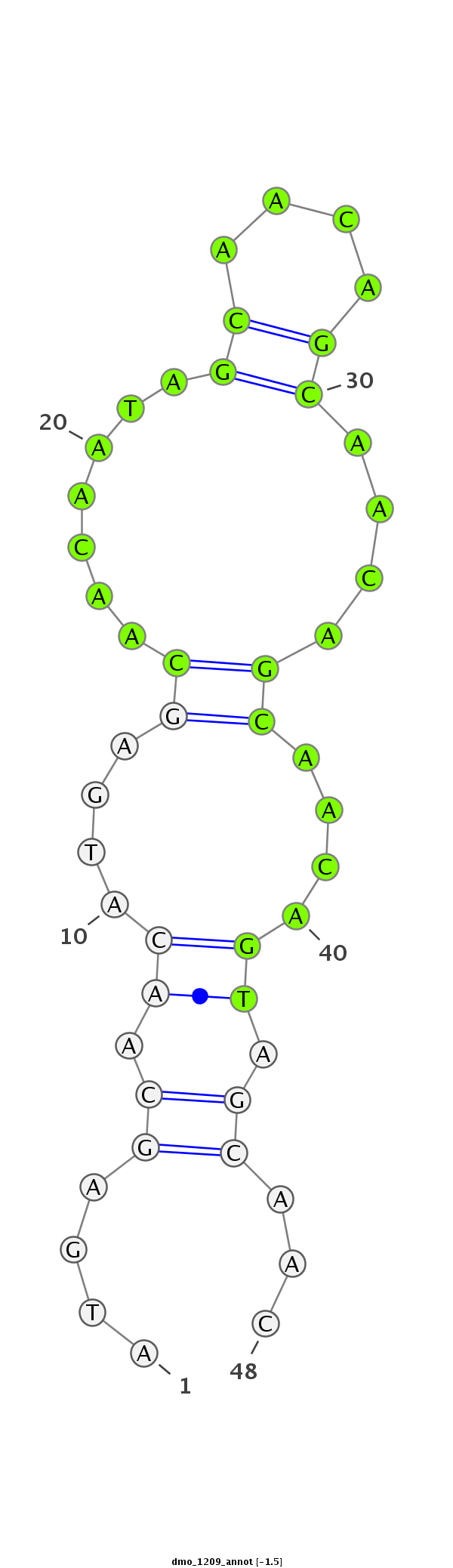
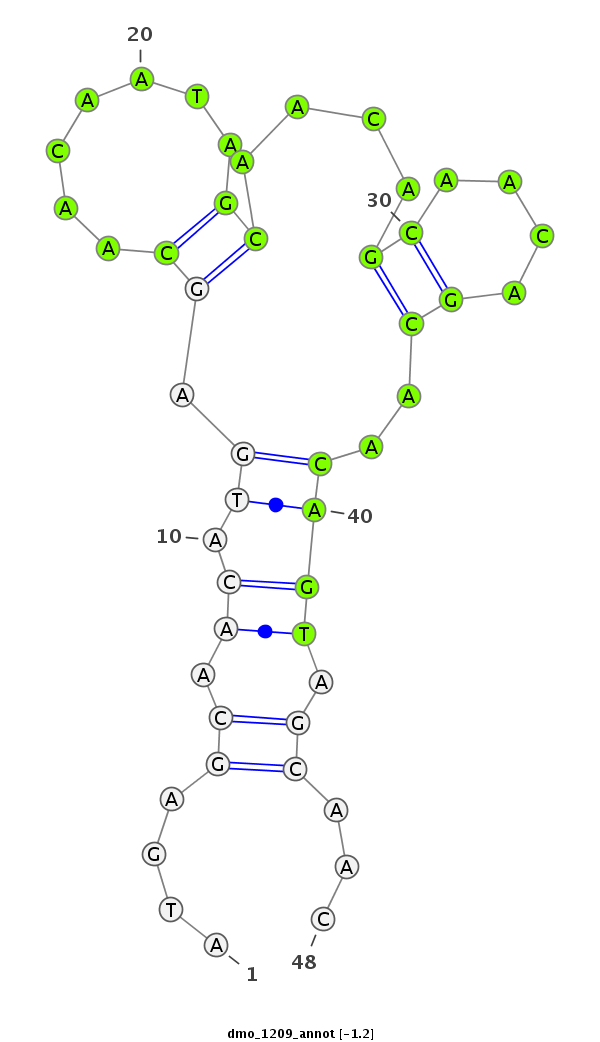
| CAGC-------AG-CA---A--CT----------------GCAACAA-----------------------CAGCAGCAGCAACAGCAACAACA--------ACAA---CA------TCAGCAGC------A---ACAGCAACAA---CAGCAA---------CAACAG---CAGCAAC---AGCAACA------ACA-----ACACG----------AGATAGAACA----------------------------------------TC--AAGAGTGCC-----C------ATGGGTGTGGCCAA--GACCGG | Size | Hit Count | Total Norm | Total | M020
Head | M045
Female-body | V117
Male-body | V118
Embryo | V119
Head |
| ............................................................................................................................................................................................................................TAGAACA----------------------------------------TC--AAGAGTGCC-----C------ATGGGTG............... | 26 | 5 | 1.00 | 5 | 0 | 1 | 0 | 4 | 0 |
| .............................................................................................................................................................................................................................AGAACA----------------------------------------TC--AAGAGTGCC-----C------ATGGG................. | 23 | 5 | 0.40 | 2 | 0 | 2 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................CG----------AGATAGAACA----------------------------------------TC--AAGAGTGCC-----C------...................... | 24 | 6 | 0.33 | 2 | 0 | 2 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................CG----------AGATAGAACA----------------------------------------TC--AAGAGTGCC-----C------A..................... | 25 | 6 | 0.33 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................TC--AAGAGTGCC-----C------ATGGGTGTGGCCA......... | 25 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................................CA----------------------------------------TC--AAGAGTGCC-----C------ATGGGTG............... | 21 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................TAGAACA----------------------------------------TC--AAGAGTGCC-----C------ATGGGT................ | 25 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................AGAACA----------------------------------------TC--AAGAGTGCC-----C------ATGGGTGTGGC........... | 29 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................................TAGAACA----------------------------------------TC--AAGAGTGCC-----C------ATGGGTGT.............. | 27 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................AGAACA----------------------------------------TC--AAGAGTGCC-----C------ATGGGT................ | 24 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................AGAACA----------------------------------------TC--AAGAGTGCC-----C------ATGG.................. | 22 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................AGATAGAACA----------------------------------------TC--AAGAGTGCC-----C------ATG................... | 25 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................GATAGAACA----------------------------------------TC--AAGAGTGCC-----C------ATG................... | 24 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................AGAACA----------------------------------------TC--AAGAGTGCC-----C------...................... | 18 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................G----------AGATAGAACA----------------------------------------TC--AAGAGTGCC-----C------A..................... | 24 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................C---AGCAACA------ACA-----ACACG----------AGAT............................................................................................. | 20 | 15 | 0.13 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................AAGAGTGCC-----C------ATGGGTGTGGC........... | 21 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................................................................................................................................................................C--AAGAGTGCC-----C------ATGGGTGTGGCCAA--T..... | 26 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................AGCAGC------A---ACAGCAACAA---CAGCAA---------........................................................................................................................................................ | 23 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ............................................................................................A--------ACAA---CA------TCAGCAGC------A---ACA................................................................................................................................................................................. | 19 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| ............................................................................................................................................ACAA---CAGCAA---------CAACAG---CAGCAA......................................................................................................................................... | 22 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................AG---CAGCAAC---AGCAACA------ACA-----ACACG----------AGA.............................................................................................. | 27 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................AACAG---CAGCAAC---AGCAACA------ACA-----ACAC............................................................................................................ | 26 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................AC---AGCAACA------ACA-----ACACG----------AGAT............................................................................................. | 21 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................AA---CA------TCAGCAGC------A---ACAGCAACAA---....................................................................................................................................................................... | 23 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................TGCC-----C------ATGGGTGTGGCCAA--GACCGG | 25 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................CC-----C------ATGGGTGTGGCCAA--GACCGG | 23 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................ACA--------ACAA---CA------TCAGCAGC------A---ACAGC............................................................................................................................................................................... | 23 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................AACAACA--------ACAA---CA------TCAGCAGC------A---AC.................................................................................................................................................................................. | 24 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................AACAACA--------ACAA---CA------TCAGCAGC------A---ACA................................................................................................................................................................................. | 25 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................CAACAA---CAGCAA---------CAACAG---CAGCA.......................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ............................................CAA-----------------------CAGCAGCAGCAACAGC.................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................CAACAACA--------ACAA---CA------TCAGCAGC------........................................................................................................................................................................................ | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................AA-----------------------CAGCAGCAGCAACAGC.................................................................................................................................................................................................................................... | 18 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................AACAA-----------------------CAGCAGCAGCAACAGC.................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................CAACAGCAACAACA--------ACAA---CA------TCAGC................................................................................................................................................................................................. | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................CAACA--------ACAA---CA------TCAGCAGC------A---AC.................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................CA--------ACAA---CA------TCAGCAGC------A---AC.................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................AGCAA---------CAACAG---CAGCAAC---AGCAAC............................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |