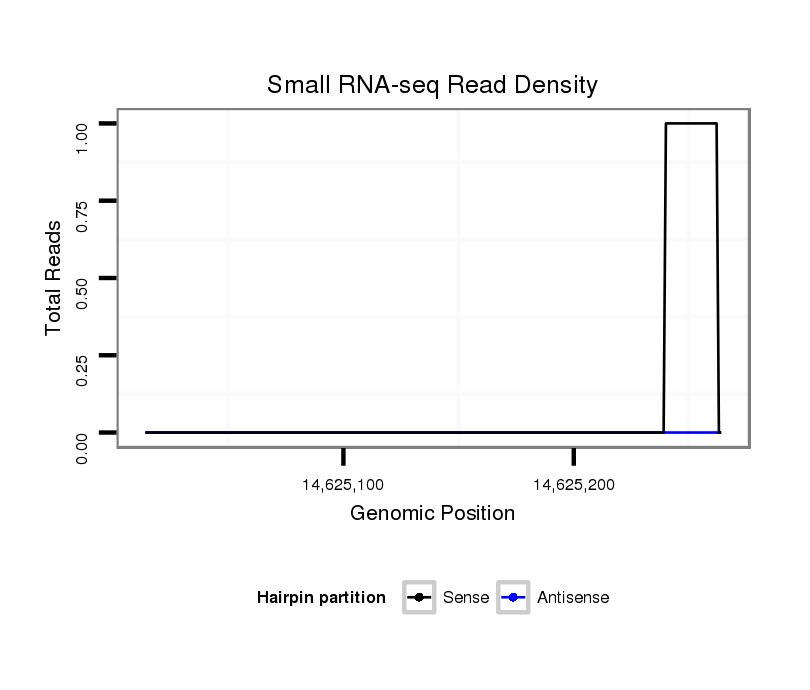
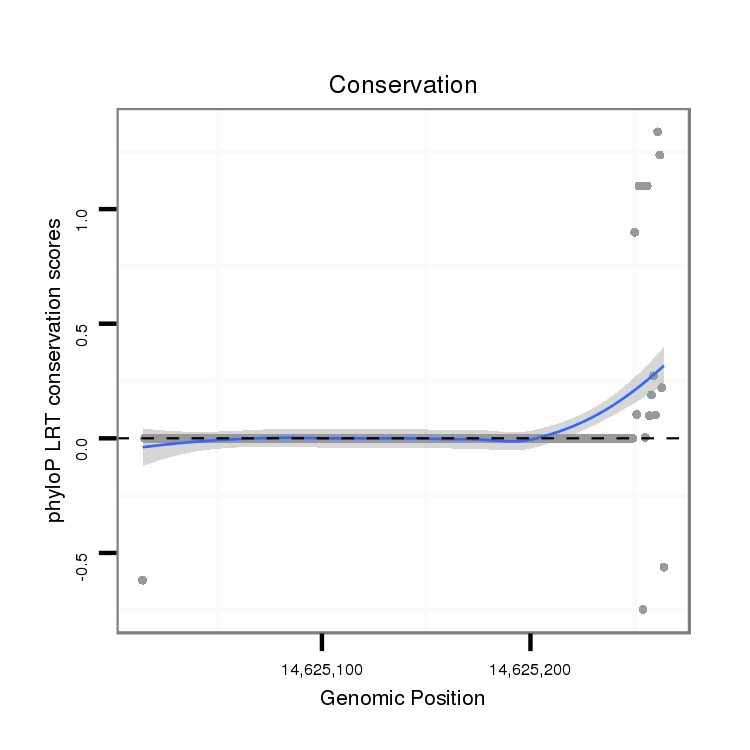
ID:dmo_1152 |
Coordinate:scaffold_6496:14625064-14625214 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dmoj\GI20671-cds]; exon [dmoj_GLEANR_5509:4]; intron [Dmoj\GI20671-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GATTTTTGCAACGTTTATCATGAGAAGTGAAACCCGTTGAAACGGCGTAATTAACTATTGATGGATTTGTTATAGAGCAAGGCATAATCGGAATACAATATGATAACTGTATTTTTTTTTAAATATATTTTTTGATACTTTTATTATGTGTATTATAATCCGTTCAATATAACATCTAATATATCTATATATTTCTTTCAGTTTTTTTCTGGATCATTCGGAGATCATAGATTATTTGCCGCCAAGATGCA **************************************************......((..((((((.(((..((((............(((((((((.((((((((..(((((....................)))))..))))))))))))).....))))............))))))).))))))..)).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V049 head |
V110 male body |
M046 female body |
V041 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ........................................AACGGCGGGAGTAACTATT................................................................................................................................................................................................ | 19 | 3 | 6 | 1.50 | 9 | 7 | 0 | 1 | 1 |
| ..................................................................................................................................................................................................................................ATAGATTATTTGCCGCCAAGATG.. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| .......................................AAACGGCGGGATTAACTAT................................................................................................................................................................................................. | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ........................................AACGGCGTGAGTAACTATG................................................................................................................................................................................................ | 19 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 |
| .......................................CAACGGCGGAAGTAACTAT................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..................................................TTGACTATTAATGGATTAGT..................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................TCAGTCTTTTTCTAGATCCT.................................. | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
CTAAAAACGTTGCAAATAGTACTCTTCACTTTGGGCAACTTTGCCGCATTAATTGATAACTACCTAAACAATATCTCGTTCCGTATTAGCCTTATGTTATACTATTGACATAAAAAAAAATTTATATAAAAAACTATGAAAATAATACACATAATATTAGGCAAGTTATATTGTAGATTATATAGATATATAAAGAAAGTCAAAAAAAGACCTAGTAAGCCTCTAGTATCTAATAAACGGCGGTTCTACGT
**************************************************......((..((((((.(((..((((............(((((((((.((((((((..(((((....................)))))..))))))))))))).....))))............))))))).))))))..)).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
|---|---|---|---|---|---|---|
| .......................CTTCGATTTGGTCAACTTTG................................................................................................................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6496:14625014-14625264 + | dmo_1152 | GATTTTTGCAACGTTTATCATGAGAAGTGAAACCCGTTGAAACGGCGTAATTAACTATTGATGGATTTGTTATAGAGCAAGGCATAATCGGAATACAATATGATAACTGTATTTTTTTTTAAATATATTTTTTGATACTTTTATTATGTGTATTATAATCCGTTCAATATAACATCTAATATATCTATATATTTCTTTCAGTTTTTTTCTGGATCATTCGGAGATCATAGATTATTTGCCGCCAAGATGCA |
| droVir3 | scaffold_12875:12247446-12247460 - | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGCCACCAAAATGCT | |
| droGri2 | scaffold_15245:12053257-12053263 - | A---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAATGC- | |
| droWil2 | scf2_1100000004513:5206270-5206283 + | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCCCTCAAATTGTC | |
| dp5 | Unknown_group_19:49694-49709 + | T-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TACCATCGCATTGCT | |
| droPer2 | scaffold_57:144744-144759 - | T-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TACCATCGCATTGCT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/17/2015 at 11:47 PM