
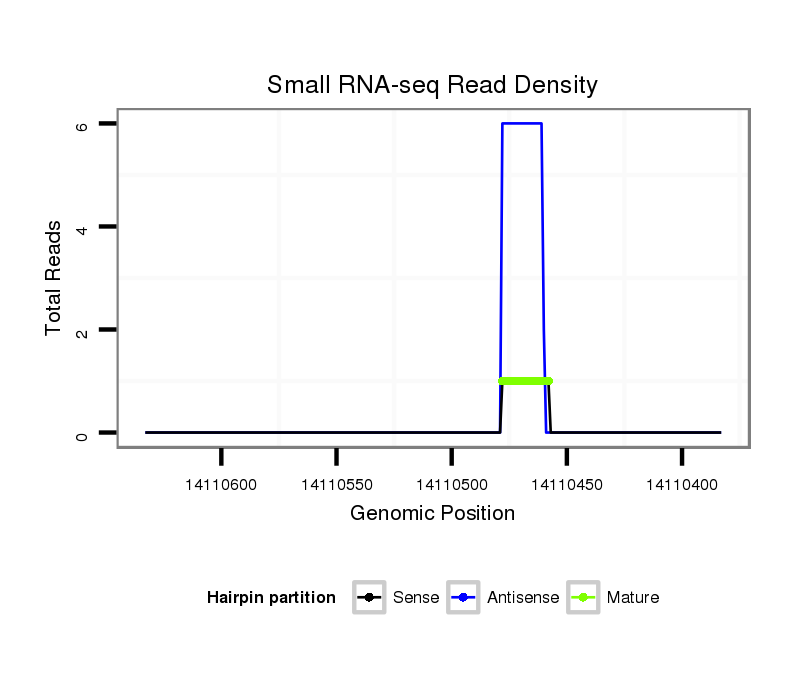
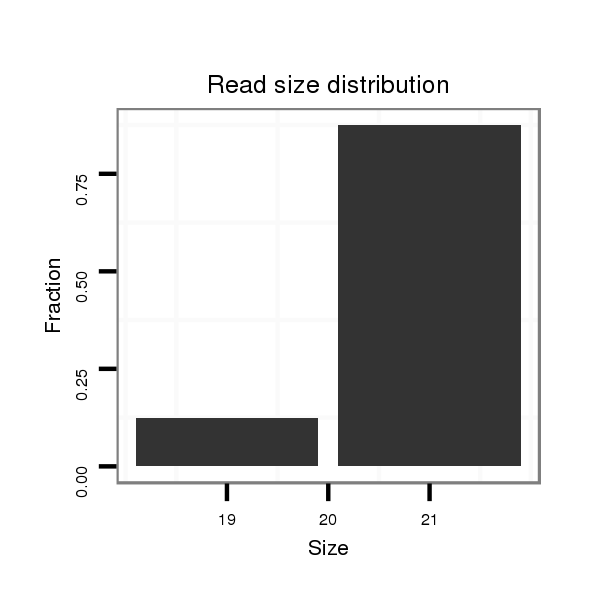
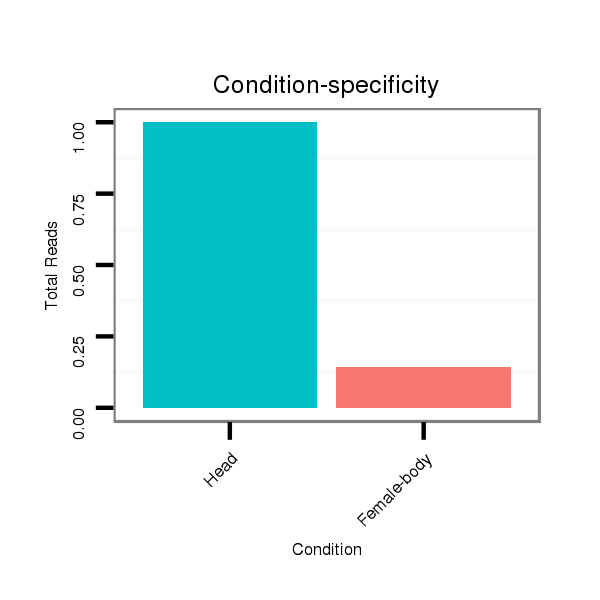
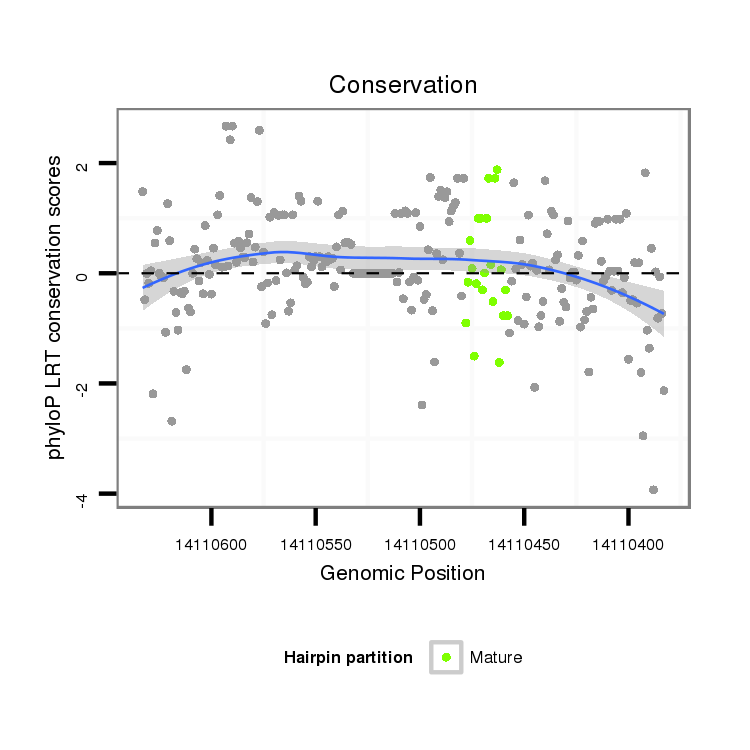
ID:dmo_1148 |
Coordinate:scaffold_6496:14110433-14110583 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -12.12 | -11.92 | -11.92 |
 |
 |
 |
exon [dmoj_GLEANR_3850:1]; CDS [Dmoj\GI18914-cds]; intron [Dmoj\GI18914-in]
| Name | Class | Family | Strand |
| (TTG)n | Simple_repeat | Simple_repeat | + |
| (ATG)n | Simple_repeat | Simple_repeat | + |
| (TTG)n | Simple_repeat | Simple_repeat | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ATCTATGAGGCGCCCACCTGGACACAATCGCTACTCAATGACCAGTCCTTGTACGTAACATCCAAGGGCGTAGGTAGCCGACATGTCGATGCTTCATTATCAACAACAAACACATCCACAACAACAACAATAACCACAACCACATCATCATCTCCATCAACATTATCATCATCATGCACAACAGCCACCAGCTCTGTCTCGACTCTAGTCTCTAGTCCAGCCTCTACCCCATCCACTCTGCCTCTGTCCAC **************************************************..............(((((...(((.((...((((..((((...........................................................................))))..)))).......)).))).)))))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
V110 male body |
V049 head |
V056 head |
|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................CTATCAACATTATCATCATCA............................................................................. | 21 | 1 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 |
| .....................................................................................................................................................ATCACTATCAACATTATCATCATCA............................................................................. | 25 | 2 | 5 | 3.80 | 19 | 19 | 0 | 0 | 0 |
| ...........................................................................................................................................................ATCAACATTATCATCATCAGTCA......................................................................... | 23 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ...........................................................................................................................................................ATCAACATTATCATCATCAGTCAT........................................................................ | 24 | 3 | 7 | 1.57 | 11 | 9 | 2 | 0 | 0 |
| .....................................................................................................................................................ATCACTATCAACATTATCATCATC.............................................................................. | 24 | 2 | 6 | 1.33 | 8 | 8 | 0 | 0 | 0 |
| ...........................................................................................................................................................ATCAACATTATCATCATCAGTC.......................................................................... | 22 | 2 | 7 | 1.14 | 8 | 4 | 4 | 0 | 0 |
| .........................................................................................................................................................CTATCAACATTATCATCATC.............................................................................. | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................ATCAACATTATCATCATCATG........................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................ATCACTATCAACATTATCATCAT............................................................................... | 23 | 2 | 11 | 0.82 | 9 | 9 | 0 | 0 | 0 |
| .............................................................................................................................................................CAACATTATCATCATCAGTCA......................................................................... | 21 | 2 | 3 | 0.67 | 2 | 0 | 1 | 0 | 1 |
| .......................................................................................................................................................CACTATCAACATTATCATCATCA............................................................................. | 23 | 2 | 12 | 0.50 | 6 | 6 | 0 | 0 | 0 |
| ....ATGAGGCGCTCAACTGGAC.................................................................................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ...........................................................................................CTTCAGTATCAACAACTAA............................................................................................................................................. | 19 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................TATCAACATTATCATCATCACG........................................................................... | 22 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................TCAACATTATCATCATCAGTC.......................................................................... | 21 | 2 | 10 | 0.30 | 3 | 1 | 2 | 0 | 0 |
| .............................................................................................................................................................CAACATTATCATCATCAGTC.......................................................................... | 20 | 2 | 14 | 0.29 | 4 | 2 | 2 | 0 | 0 |
| .............................................................................................................................................................CGACATTATCATCAGCATGC.......................................................................... | 20 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................CACTATCAACATTATCATCAT............................................................................... | 21 | 2 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 |
| ......................................................................................................................................................TCACTATCAACATTATCATCATCA............................................................................. | 24 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................ATCACTATCAACATTATCATCATCAG............................................................................ | 26 | 3 | 13 | 0.15 | 2 | 2 | 0 | 0 | 0 |
| ...........................................................................................................................................................ATCAACATTATCATCATCA............................................................................. | 19 | 0 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................ATCACTATCAACATTATCATCA................................................................................ | 22 | 2 | 14 | 0.14 | 2 | 2 | 0 | 0 | 0 |
| .........................................................................................................................................................CTATCAACATTATCATCATCAGT........................................................................... | 23 | 3 | 15 | 0.13 | 2 | 2 | 0 | 0 | 0 |
| .............................................................................................................................................................CAACATTATCATCATCAGTCAT........................................................................ | 22 | 3 | 17 | 0.12 | 2 | 2 | 0 | 0 | 0 |
| ......................................................................................................................................................TCACTATCAACATTATCATCA................................................................................ | 21 | 2 | 18 | 0.11 | 2 | 2 | 0 | 0 | 0 |
| ..............................................................................................................................................................AACATTATCATCATCAGTCA......................................................................... | 20 | 2 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................................CTTCAGTATCAACAACTAAA............................................................................................................................................ | 20 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ..........................................................................................................................................................TATCAACATTATCATCATCA............................................................................. | 20 | 1 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................TCACTATCAACATTATCATCAT............................................................................... | 22 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................TCAACATTATCATCATCAGTCAT........................................................................ | 23 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................CACTATCAACAGTATCATCATC.............................................................................. | 22 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................AACATTATCATCATCAGTC.......................................................................... | 19 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
TAGATACTCCGCGGGTGGACCTGTGTTAGCGATGAGTTACTGGTCAGGAACATGCATTGTAGGTTCCCGCATCCATCGGCTGTACAGCTACGAAGTAATAGTTGTTGTTTGTGTAGGTGTTGTTGTTGTTATTGGTGTTGGTGTAGTAGTAGAGGTAGTTGTAATAGTAGTAGTACGTGTTGTCGGTGGTCGAGACAGAGCTGAGATCAGAGATCAGGTCGGAGATGGGGTAGGTGAGACGGAGACAGGTG
**************************************************..............(((((...(((.((...((((..((((...........................................................................))))..)))).......)).))).)))))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V110 male body |
M046 female body |
V056 head |
V041 embryo |
V049 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................TAGTTGTAATAGTAGTAGTCAG.......................................................................... | 22 | 2 | 7 | 24.14 | 169 | 129 | 31 | 6 | 0 | 3 |
| ........................................................................................................................................................TGATAGTTGTAATAGTAGTAG.............................................................................. | 21 | 2 | 20 | 19.35 | 387 | 340 | 44 | 2 | 0 | 1 |
| .....................................................................................................................................................TAGTGATAGTTGTAATAGTAGTAG.............................................................................. | 24 | 2 | 6 | 12.67 | 76 | 68 | 8 | 0 | 0 | 0 |
| ...........................................................................................................................................................TAGTTGTAATAGTAGTAGTCAGT......................................................................... | 23 | 2 | 1 | 6.00 | 6 | 1 | 4 | 1 | 0 | 0 |
| .............................................................................................................................................................GTTGTAATAGTAGTAGTCAG.......................................................................... | 20 | 2 | 14 | 5.64 | 79 | 56 | 23 | 0 | 0 | 0 |
| ...........................................................................................................................................................TAGTTGTAATAGTAGTAG.............................................................................. | 18 | 0 | 8 | 4.38 | 35 | 16 | 17 | 1 | 0 | 1 |
| ..............................................................................................................................................................TTGTAATAGTAGTAGTCAG.......................................................................... | 19 | 2 | 20 | 4.00 | 80 | 43 | 36 | 1 | 0 | 0 |
| .........................................................................................................................................................GATAGTTGTAATAGTAGTAG.............................................................................. | 20 | 1 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................TAGTTGTAATAGTAGTAGT............................................................................. | 19 | 0 | 7 | 2.86 | 20 | 14 | 6 | 0 | 0 | 0 |
| .............................................................................................................................................................GTTGTAATAGTAGTAGTCAGT......................................................................... | 21 | 2 | 3 | 2.33 | 7 | 3 | 4 | 0 | 0 | 0 |
| ...........................................................................................................................................................TAGTTGTAATAGTAGTAGTC............................................................................ | 20 | 1 | 17 | 2.00 | 34 | 24 | 9 | 1 | 0 | 0 |
| .........................................................................................................................................................GATAGTTGTAATAGTAGTAGT............................................................................. | 21 | 1 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...........................................................................................................................................................TAGTTGTAATAGTAGTAGTCAGTA........................................................................ | 24 | 3 | 7 | 1.86 | 13 | 11 | 2 | 0 | 0 | 0 |
| ........................................................................................................................................................TGATAGTTGTAATAGTAGTAGT............................................................................. | 22 | 2 | 16 | 1.75 | 28 | 25 | 3 | 0 | 0 | 0 |
| ..........................................................................................................................................................ATAGTTGTAATAGTAGTAGTCAG.......................................................................... | 23 | 3 | 19 | 1.74 | 33 | 18 | 13 | 1 | 0 | 1 |
| ...........................................................................................GAAGTCATAGTTGTTGATT............................................................................................................................................. | 19 | 2 | 3 | 1.67 | 5 | 0 | 5 | 0 | 0 | 0 |
| ......................................................................................................................................................AGTGATAGTTGTAATAGTAGTAG.............................................................................. | 23 | 2 | 7 | 1.57 | 11 | 7 | 3 | 0 | 0 | 1 |
| ...........................................................................................................................................................TGGTTGTAATAGTAGTAGTCAG.......................................................................... | 22 | 3 | 17 | 1.53 | 26 | 10 | 12 | 2 | 0 | 2 |
| ..........................................................................................................................................................ATAGTTGTAATAGTAGTAG.............................................................................. | 19 | 1 | 18 | 1.50 | 27 | 18 | 9 | 0 | 0 | 0 |
| .....................................................................................................................................................TAGTGATAGTTGTAATAGTAGTAGT............................................................................. | 25 | 2 | 5 | 1.20 | 6 | 4 | 2 | 0 | 0 | 0 |
| .....................................................................................................................................................TAGTGATAGTTGTAATAGTAGTAGTCCG.......................................................................... | 28 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................AGTTGTAATAGTAGTAGTCAGT......................................................................... | 22 | 2 | 2 | 1.00 | 2 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................ATGGAGTAGGTGAGACGGAGACT.... | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................GTGATAGTTGTAATAGTAGTA............................................................................... | 21 | 2 | 20 | 0.95 | 19 | 1 | 18 | 0 | 0 | 0 |
| .........................................................................................................................................................GATAGTTGTAATAGTAGTAGTCAG.......................................................................... | 24 | 3 | 3 | 0.67 | 2 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................TAGTGATAGTTGTAATAGTAGTA............................................................................... | 23 | 2 | 11 | 0.64 | 7 | 4 | 3 | 0 | 0 | 0 |
| .....................................................................................................................................................TAGTGATAGTTGTAATAGTAGTAGTC............................................................................ | 26 | 3 | 13 | 0.62 | 8 | 8 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................TGGTTGTAATAGTAGTAG.............................................................................. | 18 | 1 | 20 | 0.60 | 12 | 3 | 8 | 1 | 0 | 0 |
| ..........................................................................................................................................................ATAGTTGTAATAGTAGTAGT............................................................................. | 20 | 1 | 13 | 0.54 | 7 | 3 | 4 | 0 | 0 | 0 |
| .............................................................................................................................................................GTTGTAATAGTAGTAGTCAGTA........................................................................ | 22 | 3 | 17 | 0.41 | 7 | 5 | 2 | 0 | 0 | 0 |
| ............................................................................................................................................................AGTTGTAATAGTAGTAGTCAG.......................................................................... | 21 | 2 | 10 | 0.40 | 4 | 2 | 2 | 0 | 0 | 0 |
| .....................................................................................................................................................TAGTGATAGTTGTAATAGTAGT................................................................................ | 22 | 2 | 14 | 0.36 | 5 | 3 | 2 | 0 | 0 | 0 |
| .......................................................................................................................................................GTGATAGTTGTAATAGTAGTAG.............................................................................. | 22 | 2 | 14 | 0.36 | 5 | 0 | 5 | 0 | 0 | 0 |
| ...........................................................................................................................................................TGGTTGTAATAGTAGTAGTCAGT......................................................................... | 23 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................TGAAGTCATAGTTGTTGATT............................................................................................................................................. | 20 | 3 | 20 | 0.30 | 6 | 2 | 4 | 0 | 0 | 0 |
| ............................................................................................AAGTCATAGTTGTTGATT............................................................................................................................................. | 18 | 2 | 11 | 0.27 | 3 | 0 | 2 | 1 | 0 | 0 |
| .......................................................................................................................................................GTGATAGTTGTAATAGTAGTAGTC............................................................................ | 24 | 3 | 20 | 0.25 | 5 | 0 | 5 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................ATGGAATTGGTGAGACGGAG....... | 20 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................AGTTGTAATAGTAGTAGTCAGTA........................................................................ | 23 | 3 | 13 | 0.23 | 3 | 2 | 1 | 0 | 0 | 0 |
| ............GGGTGGACCTATGGTATC............................................................................................................................................................................................................................. | 18 | 3 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 | 0 |
| ............................................................................................................................................................AGTTGTAATAGTAGTAGTC............................................................................ | 19 | 1 | 20 | 0.20 | 4 | 3 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................TCGAGTAGGTGAGACCGAGACA.... | 22 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................CCTGTGTTAGCGAAGACTG..................................................................................................................................................................................................................... | 19 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................GATGGTTGTAATAGTAGTAGT............................................................................. | 21 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................AGTGATAGTTGTAATAGTAGT................................................................................ | 21 | 2 | 18 | 0.17 | 3 | 1 | 2 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................GGTTGGAGATGGGGCAGGCG............... | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................CTTGTAATAGTAGTAGTCCGTA........................................................................ | 22 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................AGTGATAGTTGTAATAGTAGTAGTC............................................................................ | 25 | 3 | 16 | 0.13 | 2 | 2 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................AGTTGTAATAGTAGTAG.............................................................................. | 17 | 0 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................TTGTAATAGTAGTAGTCAGT......................................................................... | 20 | 2 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................GTGATAGTTGTAATAGTAGT................................................................................ | 20 | 2 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...........................................................................................................................................................TAGTTGTAATAGCAGTAGTCAG.......................................................................... | 22 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................TGGTAGTTGTAATAGTAGTA............................................................................... | 20 | 1 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................GTGATAGTTGTAATAGTAGTAGT............................................................................. | 23 | 2 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................CTAGTTGTAATAGTAGTAG.............................................................................. | 19 | 1 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................................................................AGTGATAGTTGTAATAGTAGTA............................................................................... | 22 | 2 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................GAAGTCATAGTTGTTGAT.............................................................................................................................................. | 18 | 2 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................GATGGTTGTAATAGTAGTA............................................................................... | 19 | 2 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................CAGTGATAGTTGTAATAGTAGTAG.............................................................................. | 24 | 3 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................TAGTTGTAATAGTAGTAGC............................................................................. | 19 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................TATTTGTAATAGTAGTAGTC............................................................................ | 20 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................TGATACTTGTAATAGTAGTAG.............................................................................. | 21 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................TGCGGTACTGGGCAGGAA......................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................GAAGTCATAGTTGTTGATTT............................................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................GGTTGTAATAGTAGTAGTCAG.......................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................TAGTGATAGTTGTAATAGCAGT................................................................................ | 22 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................GTTGTAATAGCAGTAGTCAG.......................................................................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6496:14110383-14110633 - | dmo_1148 | ATC-TATGA-G------------GCGCCCACCTGGACACA-----------------------------------------------------------------------------------------ATCGC-TAC----------------------------------------------------------------------------------------TCAATGACCAG------TCCTTGTACGTAACATCCAAGGG-----CGTAGGT-------------------------AGCC-GACATGTCGATGCTTCATTATCAACAACAAACACATCCACAACAACAACAA---TAACCACAA---CCACATCAT-------------------------------------CA-------------------------------------------TCTCCATC------AACATTATCATCATCATGCACAACAGCCACCAGCTCTGTCTCG--A-----------------------------------CTCTAGTCTCT---------------AGTCCAGCC---------------------------TCTACCCCATCC---ACT----CTGC--CTC------T---GT-CCAC |
| droVir3 | scaffold_13324:2687212-2687438 + | ATC-TATGA-G------------GCGCCCACATGGACACA-----------------------------------------------------------------------------------------ATCGC-TAG----------------------------------------------------------------------------------------TCAATGACCAG------TCCTTGTACGTACCGACG---GG-----CCAAGATGT-TGGT------TCAATGCACAGCAG----ACATGTCGATGCATCAT------------------------------------------------T-------------------------------------------------------ATCATCAACATCAACATCAACATCATTATC-------------------------------------CACCGCGACCACAAACTCTATCTTG--ATAGCT--------------------------------------------------------AGCCCAGCCTCTCCTCCCACAGTATCTACCTC-TGC-----------GCC---TGCC-TCCA--CTT------T---GT-CCAC | |
| droGri2 | scaffold_15112:4787403-4787644 - | ATC-TATGA-G------------GCGCCAACCTGGACGCA-----------------------------------------------------------------------------------------GGCGC-TAC----------------------------------------------------------------------------------------TCAATGATCAG------TCCTTGTACGTACAGACT---GG-----CGTGGGTGC-TGTTGTTCCATCAATGCACAGCAGCC-AACATGTCGATGCTTCATTATC---------------------------------AAAAT------CATCATCAT-------------------------------------CA-------------------------------------------TCTCCATC------GTCATCATCATCATTGCCTTCATCATACACAAATACTGTCTCA--ACATCG-----------------------------CAGCCAGTCTCT---------------GCTT---CC---------------------------TCTGCCCCC---CC---TGCT-TCCT--CCT------C---CT-CAAA | |
| droWil2 | scf2_1100000004822:1728313-1728557 + | ATC-TATGA-G------------GCACCACCTCT-ACACA-----------------------------------------------------------------------------------------GTCGCCATTCATCGG----------------------------------------------------------------------------------TCAATGATCAGCCGCCGCCGTTGTATGTATCATCTATG---------------TCTGGT------TCATTGAACACTAG----ACATGTGGCTACTT---------------------------CTGCTACTCCTCCAACAT------CATCATCAT-------------------------------------CA-------------------------------------------TCTTCATTATCAACAC------CATCATC---ATCGATA---GTTACTAC------T--GTCTCCTAC-GCTTCCTCTACT------------A-------ACCCAACACCAACATCAACTAC------------------------------------------TACT---ACT----ACTA--CTT------C---CA-TTAC | |
| dp5 | 3:9876147-9876308 + | ATC-TACGA-A------------GCGCCCCACCAACAA-----------------------------------------------------------------------------------------C---AC-CAGCGATCCG----------------------------------------------------------------------------------TCCAGGATCAG------TCGTTGTATGTACAGCCA---C-------------------------------------------------------------------------------------------------CAACAGCCACATCT-------GCCCCAGTTGCAGCACACACGATAGCCACTCTATCCACATCCACA------------------------TCCCATTCCAGTG------------------------------------------------------------------------------------------------------------------------------------------------------------C-----------TTC---G---T---------CCTCCTCCTCGTCGTCG | |
| droPer2 | scaffold_4:5193960-5194145 + | ATC-TACGA-A------------GCGCCCCACCAACAA-----------------------------------------------------------------------------------------C---AC-CAGCGATCCG----------------------------------------------------------------------------------TCCAGGATCAG------TCGTTGTATGTACAGCCA---C-------------------------------------------------------------------------------------------------CATCAGCCACAGCT-------GCCCCAGTTGCAGCACACACGATAGCCACTCCATCCACATCCACATCAGCATCAGCATCAGCATCAACCTCCCATTCCAGTG------------------------------------------------------------------------------------------------------------------------------------------------------------C-----------TTC---G---T---------CCTCCTCCTCGTCGTCG | |
| droAna3 | scaffold_13266:11513616-11513824 - | ATC-TACGA-G------------GCCCCCAGCCAGAGGGC-----------------------------------------------------------------------------------------GCAGC-CGG----------------------------------------------------------------------------------------CCCAGGATCAG------TTGTTGTATGTAGCCCTGAAGGGAAGCCCGGCGGC-------------------------AGAGAGGGCCGCCAACGTCTCCT----------------------------------------------------------CATCCAGT---------------ACTAGCTCATCC---------------------------------TCA----------TCTTCCTCT-----GTGATTTTCTCCACACTGC---------GCAAGTGC-GTCTCC--CCCTCG-----------------------------AATCCGATCGAG---------------AGCC--------------------------------------------CC-AA---A-GTTC--CGC------C---CT-CCAA | |
| droBip1 | scf7180000395895:56250-56451 + | AGT-CTAGT-G----------TATGGCTATCCCAGCTATCGCCGCCACTCGCAGCCACAGTTGCATCCGCAGCCGTTGTC-GCCGCATCGCTACCAGGCCAGTGTCCTGCCGCCGCCACTGCAATCCC---AGC-AAC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AG------C-------------------------------------------------------AACAACAACAACAACAACAACATCATCATCCAGCG--------------------------------------------------------------------------------------------------------CATCAG------------------------CAACATCCGCAT---CCGC-ATC-----------AGC---A---A---------CATCCGCACCA----GC | |
| droKik1 | scf7180000302698:608627-608849 + | ACC-GCGCA-G------------CAGCAACTGCATACG-------------------------------CAGCAGCATCA-GCAGCAA---------------------------------------C---AT-GTGTCAGCCG----------------------------------------------------------------------------------CCCAGCAGCAG------TTGCAACATCAA-----------------------------------------------------------------------------------------------------------CAACAG------C-------------------------------------------------------AACAGCAGCAGCAGCAGCAG----------------CATCTGCAGCAGCAG------CAGCAGC---ATCAGCA---GGTGCAGC------ATAGTCACCACCAGCAG------CAACAACATGCTGCC-------CAGCAGC---ATCAGCAGCAGCTG---CA---------TCATGAT----------C-----------CCC---A----GCAG--CAG------C---AG-CAAC | |
| droFic1 | scf7180000454044:854624-854785 + | ATC-TATGA-G------------GCACCTCAGCGATCT-----------------------------------------------------------------------------------------C---AGC-AGCAGC----------------------------------------------------------------------------------AGCAGCAGGATCAG------ATGTTGTATGTACCAACA---G-------------------------------------------------------------------------------------------------GGACTCAAAGAGACTCACCA-TCATCCAGTGCAGCCACATCGATAGCCAGTTCATCTT----------------------------------------------------------------------------------------------------------------------------CC----------------------------------------------------ACCCTAACTTCAT----------C-----------CCC---A---T---------CCTCCTCTTCTTCTCTG | |
| droEle1 | scf7180000491152:108950-109132 + | ATC-TATGA-G------------GCACCTCAGAGATCT---CAGCCGC-----------------------------------------------------------------------------AGC-------AG---------------------C-------------------------------------------AACAGCAGCAACAGCAGCAGCAGCAGCAGGATCAG------ATGTTGTATGTACCGACA---G-------------------------------------------------------------------------------------------------CAACTGAAAGAGAT-------ACATCAAGTGCAGCCTCATCGATAGCCAGTTCATCTA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCCTAACTCTCT---CCCC-ATC-----------CCC---A---T---------CCTCTCCATCTTCTCTG | |
| droRho1 | scf7180000780293:208080-208297 - | AGC-GAGGA-TCCCATACTGAA-GATTCCGTCCTTCAA-----------------------------------------------------------------------------------------G---GT-CAGTGGTCCGGCCAGCAGCAGCAGCCTGTCGCCGGGCGGACCGCTGGGTGGTGGTC---------------------------------ACCACCATCATCA--------TCCGCTGAATAACAACA---A-------------------------------------------------------------------------------------------------CAACAGCAACAGCC-------TCAGCATCAGCAACAACATCGGCAGCAGCAGCAGCAA----------------------------------------------------------------------------------CAGTCACCGGA------ACG--GCAGCA--------------------------------------------------------ATCGCA---------------------------GTC-----------CCC---A---T--------TCC---------G-CCTC | |
| droBia1 | scf7180000301754:622138-622350 - | ACT-TTG-A-G------------T--CCAACCCGGCAGCA-------------------GGTGCCTAT-CAGCAG----------------------------------------------------C---AT-CAACTGATCA-------------------------------------------GTCAGTATCAGCAAC---------------AGCAGCAGCAACAACAACAA------CAGCAGCAG--------------------------------------------------------------------------------------------------------------------------CA-------------------------------------------------------ACAACAACAACAA----------------------CAACAGCAGCAGCAG------CAGCAAC---AACAGCA---ACAGCC------ACA--GCAGCAA-------------CAACATCATCTTGCC-------CAACAGCAGCAGCACATGAGGA---------------------------------------------GTCACA---C-GCCC--CAG------A---GT-CCAC | |
| droTak1 | scf7180000415991:559085-559240 + | ATC-TATGA-G------------GCGCCTCAGAGAGCT-----------------------------------------------------------------------------------------C---AGC-AGC-------------------------------------------------------------------------------AGCAGCAGCAGCAGGATCAG------ATGTTGTATGTACCGACA---G-------------------------------------------------------------------------------------------------GAACCCAAAGAGAT-------TCATCCAGTGCAGCCACATCGATAGCCAGTTCATCTA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCCTCACTTCAT----------C-----------CCC---A---T---------CTTCCCCATCTTCTCTG | |
| droEug1 | scf7180000409663:265004-265153 - | ATC-TATGA-G------------GCGCCTCAGAGATCT-----------------------------------------------------------------------------------------C---AGC-AGC----------------------------------------------------------------------------------------AGCAGGATCAA------ATGTTGTATGTACCAACA---G-------------------------------------------------------------------------------------------------CAGCTCAAAGAGAT-------TCATCAAGTGCAGCCACATCGATAGCCAGTTCATCTA----------------------------------------------------------------------------------------------------------------------------CT----------------------------------------------------ACCCTAACTTCAT----------C-----------CCC---A---T---------CCTCCTCATCTTCTCTG | |
| dm3 | chr2R:15043373-15043531 + | ATC-TATGA-G------------GCACCACAGAGATCT-----------------------------------------------------------------------------------------C---AGC-AGC----------------------------------------------------------------------------------------AGCAGGATCAG------ATGTTGTATGTACCAACG---G-------------------------------------------------------------------------------------------------CAGCGCAAAGAGATTCATCA-TCAAGTGCTGCAGCCACATCTATAGCCAGCTCATCTA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCCTAACTTCAT---CCCC-ATC-----------GCC---A---T---------CCTCCTCTTCATCTCTG | |
| droSim2 | 2r:15693842-15693997 + | ATC-TATGA-G------------GCACCACAGAGATCT-----------------------------------------------------------------------------------------C---AGC-AGC----------------------------------------------------------------------------------------AGCAGGATCAG------ATGTTGTATGTACCAACG---G-------------------------------------------------------------------------------------------------CAACGCTAAGAGATTCA----TCATCAAGTGCAGCCACATCGATAACCAGCTCATCTA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCCTAACTTCAT---CCCC-GTC-----------CCC---A---T---------CCTCCTCTTCATCTCTG | |
| droSec2 | scaffold_15:1292674-1292851 + | ACGCGGCTA-CTACGCACTGAA--------------------------------------------------CGGCAGCTTGGAGCTA----------------------------GATTGCGACACC-------AG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAACAC------G-------------------------------------------------------AACAGCAGCAGCAGCAGCAACAACATTGGCTAC---CAACAACACCAGCAG------CAGCAGC---AACAGCA---ACCGCC-------------------------------------------------------------------------------GTTGATCCTGTCGCAGGCTCTGCTGTC---GCCATCTCC-------------GCCGCTCAA------C---GA-TCAG | |
| droYak3 | X:1068786-1068964 - | CGG-CATGGCCCAGTCGCATTAATGTCCACATCCACAT-----------------------------------------------------------------------------------------C---CAT-AGC-------------------------------------------------------------------------------------------C--------------------------------------------------------------------------------------------A---------------------------CAGCCACAGCCACAT---------CC-------------------------------------------------------ACATCCACATCCA----------------------CATCCACATCCACAT------CCACATC---CACATCC---ACATCCAC------A--TCCACATCC-ACATCCACATCC------------A-------CATCCACATCCACATCCACATC------------------------------------------CACA---TCC----ACAT--CCA------C---GT-CCAC | |
| droEre2 | scaffold_4845:9243317-9243466 + | ATC-TACGA-G------------GCACCGCAGAGATCT-----------------------------------------------------------------------------------------CAGCAGC-AGC----------------------------------------------------------------------------------------AGCAGGATCAG------ATGTTGTATGTACCAACG---G-------------------------------------------------------------------------------------------------CAACTCAAAGAGAT-------TCACCAAGTGCAGCCACATCGATAGCCAGTTCATCTA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCCTAACTTCGT----------C-----------CCC---A---T---------CCTCCTCTTCATCTCTG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/17/2015 at 11:46 PM