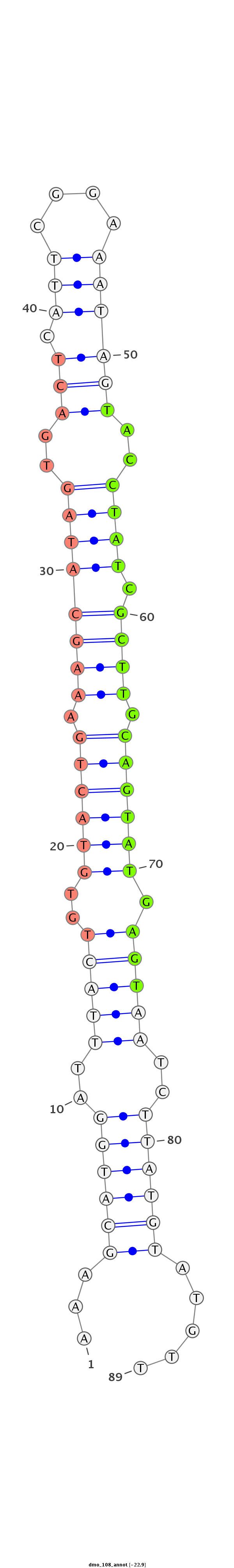
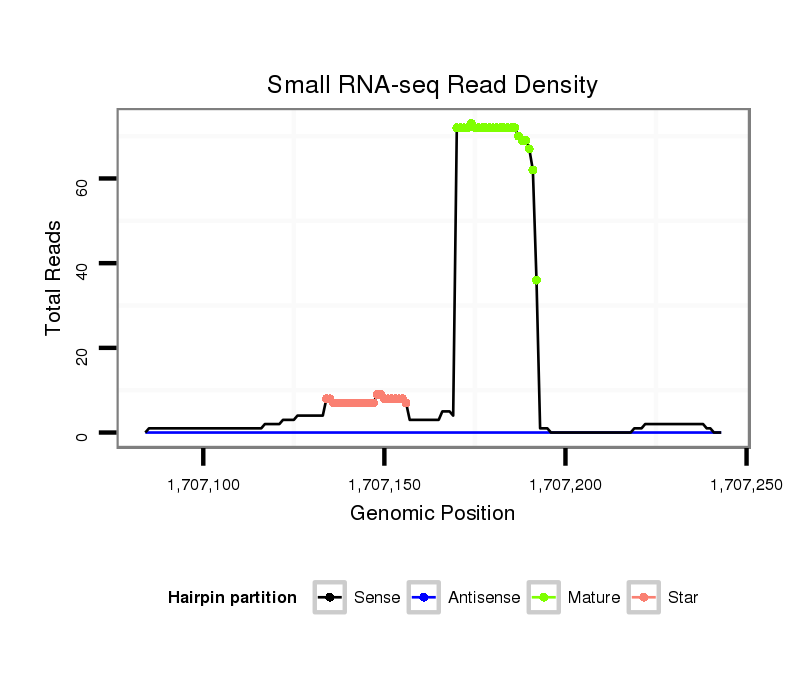
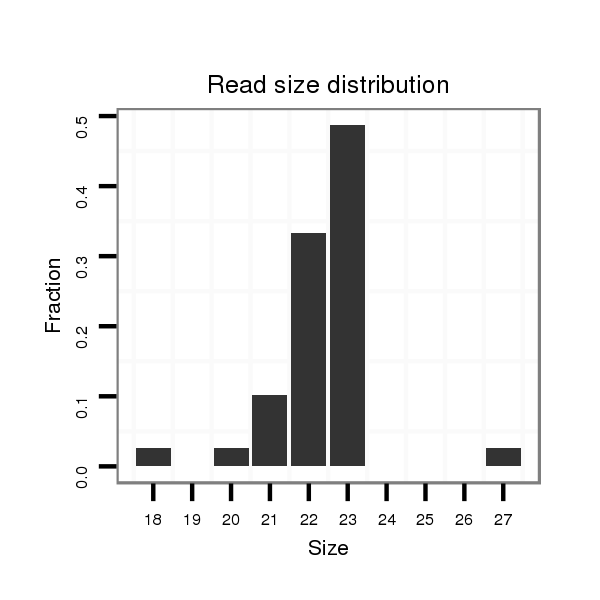
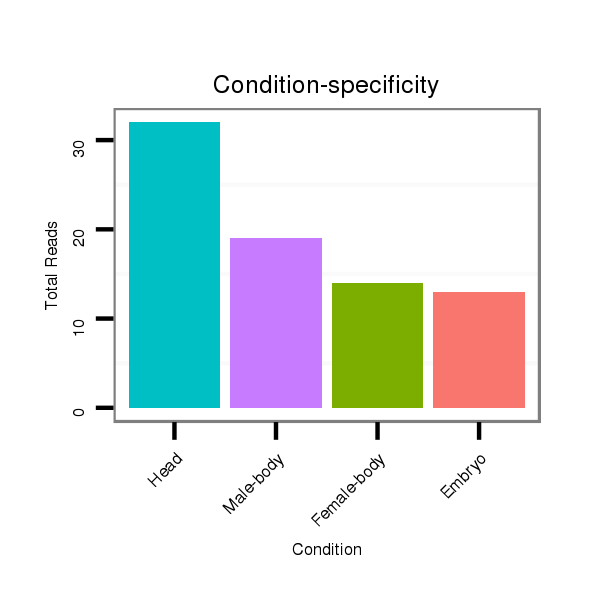
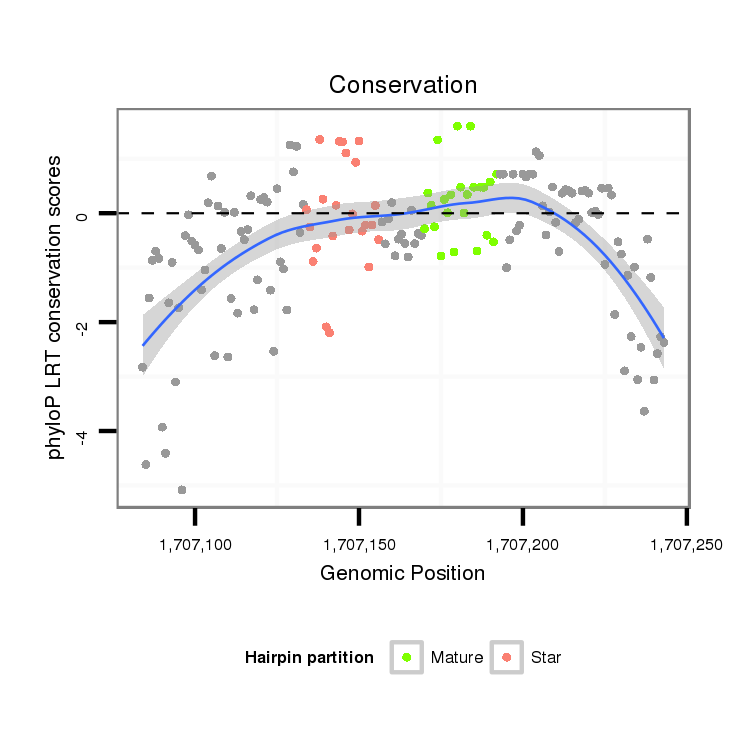
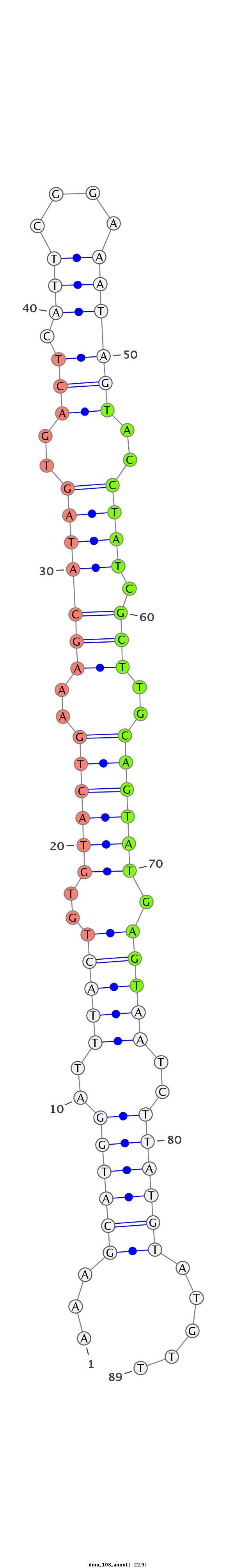
ID:dmo_108 |
Coordinate:scaffold_6541:1707134-1707193 + |
Confidence:confident |
Class:Canonical miRNA |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -22.9 | -22.7 | -22.7 |
 |
 |
 |
intergenic
No Repeatable elements found
|
TGATTTACTTATCAAATGAGTTGATCTATGTGGATAAAGCATGGATTTACTGTGTACTGAAAGCATAGTGACTCATTCGGAAATAGTACCTATCGCTTGCAGTATGAGTAATCTTATGTATGTTTTATCTTCAGTTGCAACTCCTACAAATGCAACCAAA
***********************************...((((((..(((((..((((((.((((((((..(((.(((....))))))..)))).)))).)))))).)))))..)))))).....************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | V110 male body |
V056 head |
M046 female body |
V049 head |
M060 embryo |
V041 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................TACCTATCGCTTGCAGTATGAGT................................................... | 23 | 0 | 1 | 35.00 | 35 | 9 | 10 | 5 | 5 | 3 | 3 |
| ......................................................................................TACCTATCGCTTGCAGTATGAG.................................................... | 22 | 0 | 1 | 25.00 | 25 | 6 | 5 | 4 | 4 | 2 | 4 |
| ......................................................................................TACCTATCGCTTGCAGTATGA..................................................... | 21 | 0 | 1 | 5.00 | 5 | 0 | 2 | 0 | 3 | 0 | 0 |
| ......................................................................................TACCTATCGCTTGCAGTATGAGTT.................................................. | 24 | 1 | 1 | 4.00 | 4 | 2 | 1 | 0 | 0 | 1 | 0 |
| ..................................................TGTGTACTGAAAGCATAGTGACT....................................................................................... | 23 | 0 | 1 | 3.00 | 3 | 1 | 0 | 1 | 1 | 0 | 0 |
| ..................................................................................ATAGTACCTATCGCTTGCAGT......................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| ......................................................................................TACCTATCGCTTGCAGTATG...................................................... | 20 | 0 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 |
| ................................................................ATAGTGACTCATTCGGAAATAGTACCT..................................................................... | 27 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TACCTATCGCTTGCAGTATGAGA................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................ATTACCTATCGCTTGCAGTATGAG.................................................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................TACCTATCGCTTGAAGTATG...................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................ATAGTACCTACCCCTTGCAGT......................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................ATAAAGCATGGATTTACTG............................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................TATCGCTTGCAGTATGAG.................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................TACCTATCGCTTGCAGTATGAGC................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TACTTATCGCCTGCAGTATGAGT................................................... | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................ATCGCTTGCAGTATGAGTAAT................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................TGCCTATCGCTTGCAGTATGAGT................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................TACCTATCGCTTGCAGTA........................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................CACCTATCGCTTGCAGCATGAGT................................................... | 23 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................TGCCTATCGCTTGCAGTATGAG.................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................TACCTATCGCTTTCAGTATGAGT................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TGTGTACTGAAAGCATAGTGACTT...................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................GCATGGATTTACTGTGTACTGAAAGCAT.............................................................................................. | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................GGATTTACTGTGTACTGAAAGC................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................TGCAACTCCTACAAATGCAA..... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................AACTCCTACAAATGCAACC... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................TACCTATCGCTTGCAGTATGCG.................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................TACCTATCGCTTGAAGTATGAGT................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................TACCTATCGCTTGCAGTATGAA.................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................CTATGTGGATAAAGCATGG.................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................ATAGTGACTCATTCGGAAATA........................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TGTGTACTGAAAGCATAGTGAC........................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................TACCTATCGCTTGCAGTATGAGTCA................................................. | 25 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................TACCTATCGCTTGCAGCATGAGTA.................................................. | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................TTACTGTGTACTGAAAGCATGGTGAC........................................................................................ | 26 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................ATTTACTGTGTACTGAAAGCATAGTGACT....................................................................................... | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .GATTTACTTATCAAATGAGTTGAT....................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....TTACTTATCAAATGAGTTG......................................................................................................................................... | 19 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................TCCCTATCGATTGCAGTATG...................................................... | 20 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................ATCTATGTTGAGAAAGCA....................................................................................................................... | 18 | 2 | 9 | 0.33 | 3 | 0 | 2 | 0 | 1 | 0 | 0 |
| ......................................................................................TACCTATCTCTTGCAGTATCTG.................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
|
ACTAAATGAATAGTTTACTCAACTAGATACACCTATTTCGTACCTAAATGACACATGACTTTCGTATCACTGAGTAAGCCTTTATCATGGATAGCGAACGTCATACTCATTAGAATACATACAAAATAGAAGTCAACGTTGAGGATGTTTACGTTGGTTT
************************************...((((((..(((((..((((((.((((((((..(((.(((....))))))..)))).)))).)))))).)))))..)))))).....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V056 head |
V049 head |
|---|---|---|---|---|---|---|---|
| .......................................................................GAGGACGCCTTTATCATGGATA................................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 |
| .......................................................................GAGTAAGCCGTGATCATGAATA................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 1 |
| .......................................................................................TGGAAAGCGAATGTCATAAT..................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 1 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droMoj3 | scaffold_6541:1707084-1707243 + | dmo_108 | confident | TGAT--------------------TTACTTA-------------------------------------------------------------------TCAAATGAGTT-----------GATCT-ATGTGGATAAA----------------------------------------------------------------G------------------------------CATGG--------------------------ATTTAC------TGTGTACT----------------------GAAAGCATAGTGACTCATTCGGAAATAGTACCTATCGCTTGCAGTATGAGTAATCTTATGTATGTTTT-------------------------------------------------------------------------------------ATCTTCAGTTG---CAACTCCTA--CAA------------------------------------------------------ATGCAA-----------------------------CCAAA |
| droVir3 | scaffold_13036:588437-588541 + | ACCG--------------------ACAGTAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACCAGCAGTCTCTGTCAGAGTACCTTCTGCTTGCAATATGGGTAAGCTGGTGTATGTAAT-------------------------------------------------------------------------------------ATCTTCAGTGG---CAACACCTA--CAA------------------------------------------------------CTCCGA-----------------------------CCCTA | ||
| droGri2 | scaffold_15203:4419951-4420034 - | TCTG--------------------TAACC-------------------------------------------------------------------------------------G--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGCCTATTGTTTGCAAAATGAATAAAGTGGTGTATGTATT-------------------------------------------------------------------------------------TACCTCAGTTG---CATCGCCGG--TTG------------------------------------------------------GTGCAA-----------------------------CAAAA | ||
| droWil2 | scf2_1100000004521:1182334-1182479 - | GAAT--------------------CTGTTCG-------------------------------------------------------------------TCAAAAACGTT-----------GGTCA-A-----------ATATAAA-----------------------------------------------------------------------------------------T-----------------------------------------------------------------------------------------------------------------------------------------CAGTATT-CCTACCACAAAATTGGTTGAACATTCATTCTGAAATAGTTTTTGTCTTTTATGGAAGTCATCTAACAACCA------------------------------AATCCTCAACTGCAAT-TACTA-------------------------------------TTACAA-----------------------------GCAAG | ||
| dp5 | Unknown_group_103:1143-1237 - | AAAT--------------------ATTGTCT-------------------------------------------------------------------TTGATA----------GGAGCTGA-AGCTTCAGAAGAAATATACG--------------------------------------------------------------------------------------------AAT----------------------------TGTTCCG------TACA----------------------GTAAG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CATTCTAAATCT------T-AAAGATTTATCCAGAAAT | ||
| droPer2 | scaffold_2246:2162-2333 - | TTAT--------------------TAATTTA----------------------------------------------------ATTTATTTATATTGGCGTGGACT--------GGGCCCAT-AA--------------------------------------------------------------------------------------------------------------------------------------------------------CCTATTGGTAGACTAGCTATGACAATTAGTA-----AACGGAAGACAGTACCAAGTTATGTTTTTCTTTAAAGATTAT----------------------------------------------------------------TTATGTATTTCTTGATT----------------------ACTTTTTGCTG---CAACACCGG--TCC------------------------------------------------------ACACAC--------------T-TG----------AGATCA | ||
| droAna3 | scaffold_13334:685267-685317 + | AGCC--------------------TTTTGTT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGCTTT-------------------------------------------------------------------------------------TTCTTCTATTG---ATTCTCTTG--AAA------------------------------------------------------ATAAAA-----------------------------ACAAA | ||
| droBip1 | scf7180000395142:32109-32178 - | TAAT--------------------TTATTTC-TAGTTCTAAGAATAAGGCATTTGAGGCATACAAAAACATAATATT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAAAAA-----------------------------ATAAT | ||
| droKik1 | scf7180000302590:78777-78933 + | TTCTTTAAA------------------TATC-CAGGCGAGTGAATCAAGCC------------------------------------------------------ATTT-----------TATAT-AAAT--------CTATA--AGCTGGGGTTTATTTTAAATAGAGCATGGTTTCAAAACTCATCAAGGGAAATCCAAATAATTTTTTTCATTATTAAAATGGAAATAT-------------------------------------------------------------------------------------------------------------------------AA----------------------------------------------------------------------TTATATT------------------------------------------------------------------------------------------------A--TATA-TTT--------------------------------------TTT | ||
| droFic1 | scf7180000453769:5842-5916 - | AATT--------------------TGGGG---------------------------------------------------------------------TCAAATGGGGT-----------CAAAA-AATAGAAAAAA----------------------------------------------------------------G------------------------------AATGAA----------------------TGGAT------CGGTTAAGTTCT----------------------AAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAA-----------------------------------------------TA-G--------------------------------------GTACT | ||
| droEle1 | scf7180000490489:11078-11207 - | CTGTACCATTGACTTACCAAATTGAAAATA---------ATGCAGTACAAG-------T----------------------------------GTTAAAAAATTGTGTTCGAACGT----------GTTAAAAACAC----------------------------------------------------------------GAAATTTTATACATTGC----------------------------------------------T---------------------------------------------------------------------------TGCTTAAAATACGAA----------------------------------------------------------------------ATATTTT------------------------------------------------------------------------------------------------AAGTGTC-TTT--------------------------------------TAT | ||
| droRho1 | scf7180000759908:15346-15501 - | TAAC--------------------TTATTTTACAGACTTTTGAATAATGAT---------------------------------------------TTCTAA---------------------------------------------------ATCATTTTATT---------------------------------------------------------------------------------------------------------------------------------------------TAAAGCTTAATTACCGATTGATAACTGTTATTTCTTGTTTAAAATATCAA----------------------------------------------------------------------ATATAA------------AATTGAGAAAATCTTTACTCCGCTTATGATTTCTCTCA--GTG------------------------------------------------------CACACA-----------------------------CCCAA | ||
| droBia1 | scf7180000302433:9753-9865 - | ATAT--------------------TTATATA-------------------------------------------------------------------TCAAAAGAATT-----------CATAT-ATATAG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAAAATTATTTATTACA----------------------------------------------------------------------------------------TTAA--TGAGTCGAACTAATGTCCCGTCATTTAAATTTTGATTTCACTTATTTAAGTTTTAG--------------------------------------TTT | ||
| droTak1 | scf7180000415941:338694-338823 + | ATAT--------------------TTCAAAA---------------A---------------------TGAAATATAAATTAAGTTTATTAATGTTTGTTTGATGTGG-------------------------------------------------------------------------------------------------------------------------------------------------------------TTAT------TGTGTAGG----------------------TAAAGTTTAATAACCGCTTCAAA--TTGCA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTTTATAAATCTGATTTTTACAAATTTTATAAATGTTT | ||
| droEug1 | scf7180000409027:17786-17905 + | TAAT--------------------TATTTTG-------------------------------------------------------------------TGTAATATTTT-----------AAAG-----------------------------------------------------------------------------------------ATAAG-----TGGAAAAAAAATTAATTTATCTCTTTTGGCACAAGTTTAAGT---------------------------------------------------------------------------CGGTAAAAACGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------AGAGTAGTTTAAAGTATAGC-TTATCCTCGG-----------------------------CTGTA | ||
| dm3 | chr4:769657-769732 - | TAAT--------------------CTTAATA---------------T---------------------------------------------TGTTTGCCAAATGTGTT-----------TAAAT-GTTAAGCCAAA----------------------------------------------------------------G------------------------------CAGGT--------------------------T----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATA---GC--TGGACTGA--------------------------------------------------ACTGA-----------------------------TAATG | ||
| droSim2 | 2l:21033289-21033441 - | TGGC--------------------TTACTTA-------------------------------------------------------------------AG-------------------------ACTAAGGAGCCA-ATACAAAAA--------TTTCCAAAGCAGAAAATAGTTTTGAAATGTGTAA-------------------------------------------ACTGAA----------------------------TGTACCTGGCA----------------------------------------------------------------------------------------------------------AATAGT------------------------------------------------------------CC------------------------------AATCAAAAACATTAACGTGTTATCATCGGCAGTTGGAGTATATTACTATGTATT-TTA--------------------------------------TCT | ||
| droSec2 | scaffold_28:298602-298755 - | TGGC--------------------TTACTTA-------------------------------------------------------------------AG-------------------------ACTAAGGAGCCAAATACAAAAA--------TTTCCAATGCAGAAAATAGTTTTGAAATGTGTAA-------------------------------------------ACTGAA----------------------------TGTACCTGGCA----------------------------------------------------------------------------------------------------------AATAGT------------------------------------------------------------CC------------------------------AATCAAAAACATTAACGTGTTATCATCGGCAGTTGGAGTATATTACTATGTATT-TTA--------------------------------------TCT | ||
| droYak3 | 2R:10750491-10750547 + | AAAA--------------------TTTAAAA-------------------------------------------------------------------TTATAAAT-----------------------------------------------------------------------------------------------------------ATTTT-----TGGGAATAAAGTAG--------------------------------------------------------------------------------------------------------------------AT----------------------------------------------------------------------TTAATT-------------------------------------------------------------------------------------------------------------TGAGTG-----------------------------CTAAA | ||
| droEre2 | scaffold_4784:23832881-23832949 - | AGAA--------------------CTGCTTT-------------------------------------------------------------------TCAACAGACTA-------------TAA----------------------------TTTATTTTGGTTGAA---------------------------------G------------------------------CT--------------------------TGCCT------TGGTCAAATA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAAAAA-----------------------------CCATA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 10/20/2015 at 07:01 PM