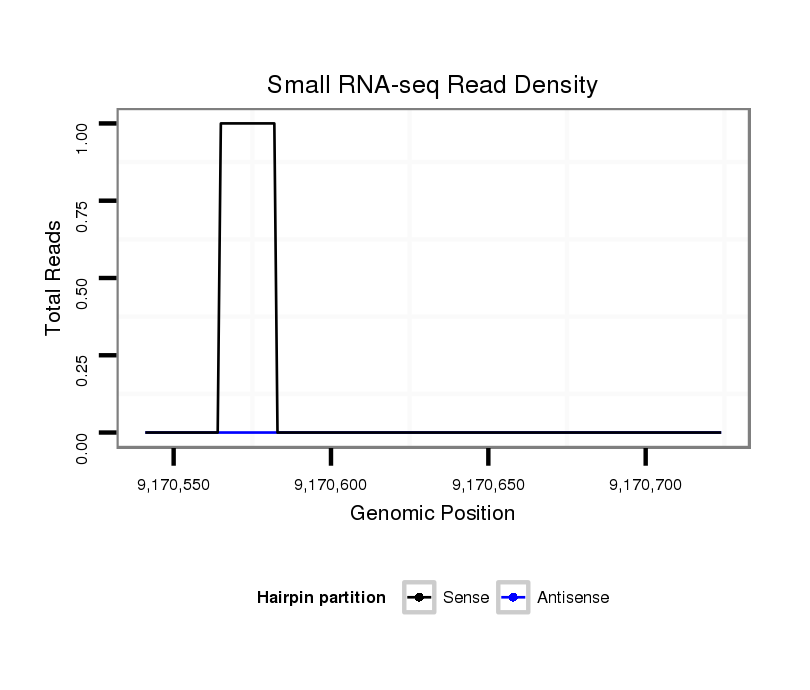
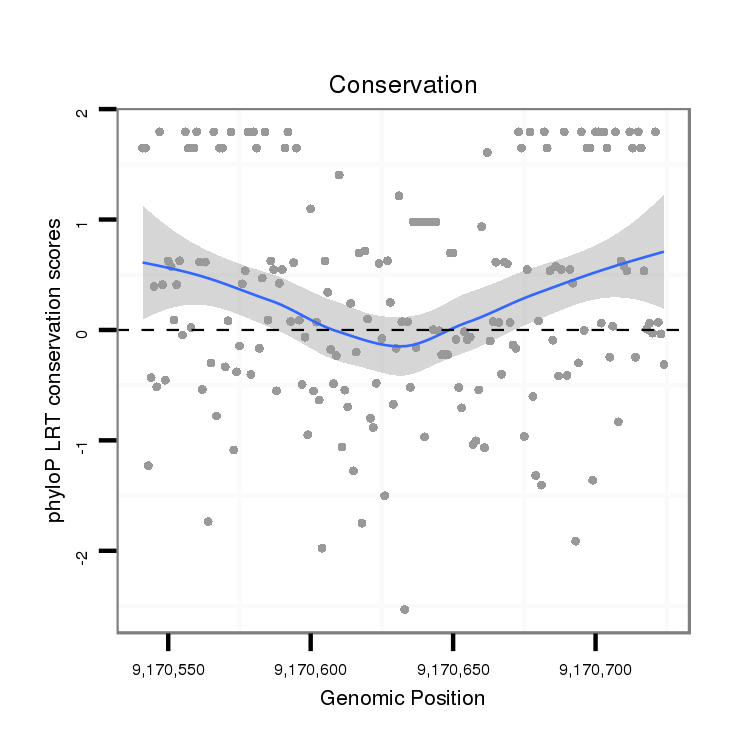
ID:dmo_1051 |
Coordinate:scaffold_6496:9170591-9170674 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dmoj\GI20304-cds]; exon [dmoj_GLEANR_5171:3]; exon [dmoj_GLEANR_5171:2]; CDS [Dmoj\GI20304-cds]; intron [Dmoj\GI20304-in]
No Repeatable elements found
| ##################################################------------------------------------------------------------------------------------################################################## CGCTCCAATAAAAACACGCATCTTATTGCACTCGTACTCTGCGTAACGGGGTGAGTTAGCTGATATATGATATATCCATACTATACATTTATATCTCATCTATATTTTCCCTTGCTCTCTCTTCTTCTTTCCAGCCTGATTTGCTTGGTTCCCTTGCCATATTGCCTCAAGAGCTGCCGCAGCC **************************************************..((((.((..((((((.((((((...............)))))).....))))))....)).)))).................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M046 female body |
M060 embryo |
V049 head |
|---|---|---|---|---|---|---|---|---|
| ........................ATTGCACTCGTACTCTGC.............................................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..................................................................ATGATATATCCATTCTCTACT................................................................................................. | 21 | 3 | 10 | 0.50 | 5 | 5 | 0 | 0 |
| ................................................................................................................................TTACAGACTGATTTGATTGGTT.................................. | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 |
| ...........................................................................................................TCCCCTGCTCTCCCTTCTTC......................................................... | 20 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 |
| ..................................................................ATGATATATCCATTCTCTA................................................................................................... | 19 | 2 | 12 | 0.08 | 1 | 1 | 0 | 0 |
| ...............................................................................................................................TTTCCATCCTGTTTTGCT....................................... | 18 | 2 | 18 | 0.06 | 1 | 1 | 0 | 0 |
|
GCGAGGTTATTTTTGTGCGTAGAATAACGTGAGCATGAGACGCATTGCCCCACTCAATCGACTATATACTATATAGGTATGATATGTAAATATAGAGTAGATATAAAAGGGAACGAGAGAGAAGAAGAAAGGTCGGACTAAACGAACCAAGGGAACGGTATAACGGAGTTCTCGACGGCGTCGG
**************************************************..((((.((..((((((.((((((...............)))))).....))))))....)).)))).................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V110 male body |
M046 female body |
M060 embryo |
V056 head |
|---|---|---|---|---|---|---|---|---|---|
| ..................................................................TACTATATAGGTAAGAGATG.................................................................................................. | 20 | 2 | 3 | 6.33 | 19 | 15 | 4 | 0 | 0 |
| ........................................CCCGTTGCCCCACTCGATCGACT......................................................................................................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ..................................................................TACTATATAGGTAAGAGATGA................................................................................................. | 21 | 3 | 10 | 0.50 | 5 | 4 | 1 | 0 | 0 |
| ..................................................................TACTATATAGGTAAGAGAT................................................................................................... | 19 | 2 | 12 | 0.33 | 4 | 0 | 4 | 0 | 0 |
| ............................................................................................................................AAGAAAGGGGCGACTAAACG........................................ | 20 | 3 | 12 | 0.17 | 2 | 2 | 0 | 0 | 0 |
| ...............................................................................................................................AAAGGTAGGACAAAACGA....................................... | 18 | 2 | 18 | 0.17 | 3 | 0 | 3 | 0 | 0 |
| ....................................................................................................................GAGAGAAGAAGAAAGGTATGAT.............................................. | 22 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 |
| .................................................................TTACTATATAGGTAAGAGATG.................................................................................................. | 21 | 3 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................GAGAGAAGAAGAAAGGTAAGAT.............................................. | 22 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droMoj3 | scaffold_6496:9170541-9170724 + | dmo_1051 | CGCTCCAATAAAAACACGCATCTTATTGCACTCGTACTCTGCGTAACGGGGTGAGTTAG--C---TGA-------------T------------ATATGATATA----TCCATACTATACATTT-ATATCTCATCTAT---AT------------------------TTTCC----------CTTGC-----------TCTC--TCTTCTTCTTTCCAGCCTGATTTGCTTGGTTCCCTTGCCATATTGCCTCAAGAGCTGCCGCAGCC |
| droVir3 | scaffold_12875:13751363-13751538 + | CGTGCGAATAAGAACACGCACCTGTTGGCCCTTATACTCTGCCTAACCGGGTGAGTCAG--CTAACGTCAGCAGCTGCTGGC------------ATGGAACTAAATTGC----CCTGGTGACGCC-------------------------------------------------------------------------CTTC--TGTTCTGTTTTCCAGGCTGGTCTGCCTGGTGCCGCTCCCGTATTGCCTCAAGAGCTGCCGCAGCT | |
| droGri2 | scaffold_15245:17491476-17491652 - | CGAGCGAACAAAAACACGCATATACTAGCCCTCCTTCTCTGTCTAACTGGGTGAGCCGC------TGG-------------T------------GT-----------------TGCATACATTTTATGTATTATGTAT---ATCCGGAATTTGTACAAT---------------------------------------CGGAAT--TTCTTTTGTTTAGGCTATGGTGCTTTATGCCACTACCCTACTGTCTAAAGAGTTGCCGCAGTT | |
| droWil2 | scf2_1100000004513:4065133-4065320 - | CGTCCCAATTCGAGAACTCATCTAATTGCTTTGATTCTGTGTCTGTTTCAGTAAGTAAAAG----TGTTGACAATTTCCAAT------------A-------------G----TGACTAAG------------------------------------TTAATCACACCCCCC----------CCCACTCCCTCCCCCTCGCATT--TGCTTCTTTTTAGATTATATTGCTGTGTTTGTTTGCCATATTGCATCTCGAGTTGCATGAACA | |
| dp5 | 3:4031298-4031458 + | CGAGTGACGAAGCACACCCATGCCTTTGCCTTTGCTCTCTGTCTGACGGGGTGAGTCTCGGCCATCGG-------------C------------AGGGGATGCC----GTCATTCCATTCATTCCACCC-------------------------------------------------------------------------------TCTGCTCTTAGCCTTCTCTGCCTGGTGCCGCTGCCCTACTGTCTGAAGAGCTGCCAGAGCG | |
| droPer2 | scaffold_2:4217363-4217523 + | CGAGTGACGAAGCACACCCATGCCTTTGCCTTTGCTCTCTGTCTGACGGGGTGAGTCTCGGCCATCGG-------------T------------AGGGGATGCC----GTCTTTCCATTCATTCCACAC-------------------------------------------------------------------------------TCTGCTCTTAGCCTTCTCTGCCTGGTGCCGCTGCCCTACTGTCTGAAGAGCTGCCAGAGCG | |
| droAna3 | scaffold_13266:10905985-10906141 - | CGAATAACCAAGCAAACCCATTTGTTTGCACTGGCTTTATGCCTAACGGGGTATGCTTT------TTG-------------A------------AGAAAACTCAAATAA----TTTTTTAA------------------------------------AC---------------------------------------ATTTAT--TATTTTTTTTCAGACTTATATGTCTTGTTCCCTTGCCGTATTGCCTAAAAAGCTGTCAAAGTG | |
| droKik1 | scf7180000302470:449263-449498 - | CGCGTGACCAAGCAAACCCATCTTTTCGCCCTTGGTCTTTGCCTAACGGGGTGAGTTTTTCCCATAA---ATTGCTGGAAACATGACACACAAAAGGAAAGTTAATTGT----TTAATTGATGCCATTTATCATTTACGAAAACCCGATTTTGTTTACA-------------TTAAAAAACAAAGGA-----------CTGA--TATTCTATTTCGCAGACTATTATGCCTGGTGCCTTTGCCGTATTGTTTGAAGAGTTGCAAGAATG |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
Generated: 05/17/2015 at 11:35 PM