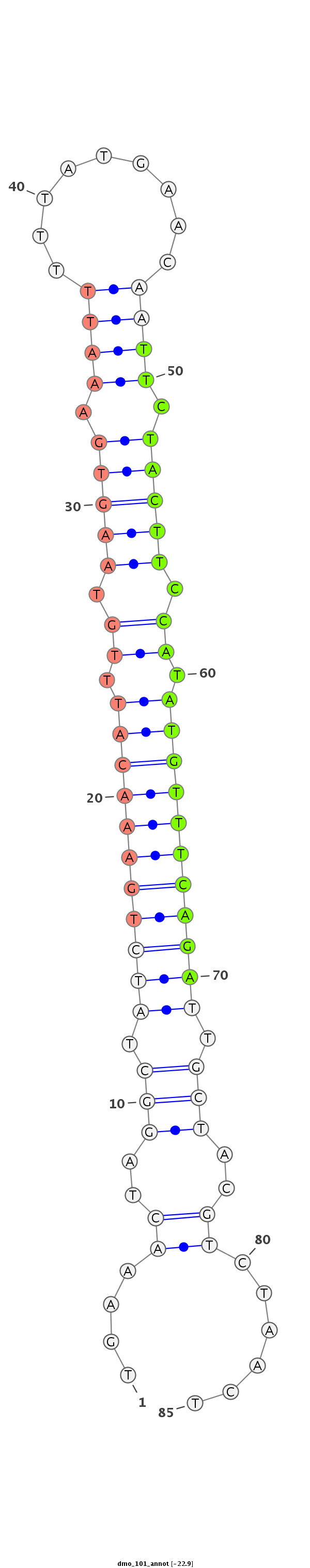
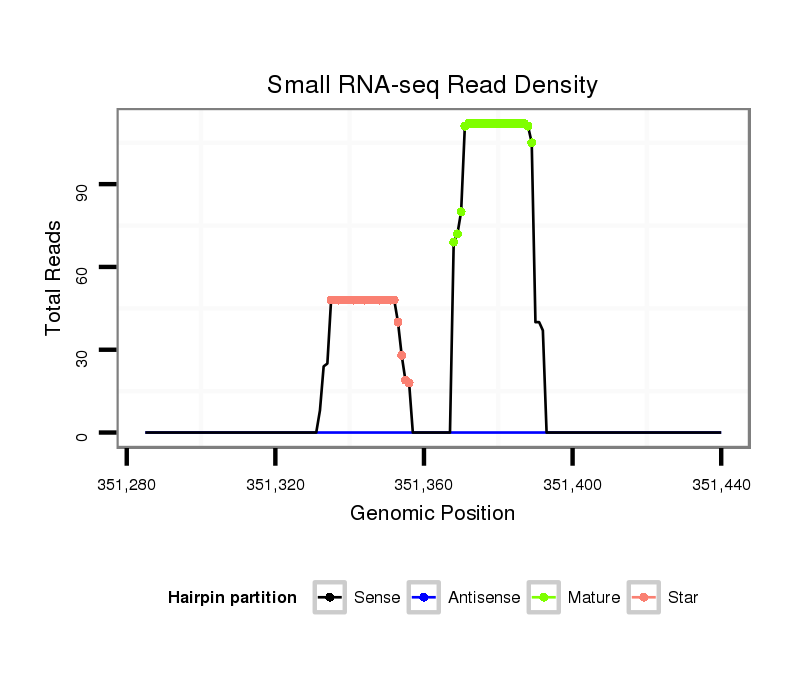
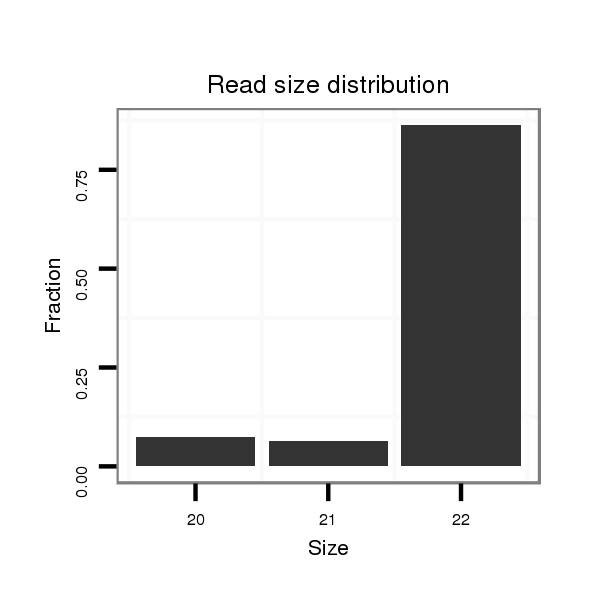


ID:dmo_101 |
Coordinate:scaffold_6328:351335-351390 + |
Confidence:confident |
Class:Canonical miRNA |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
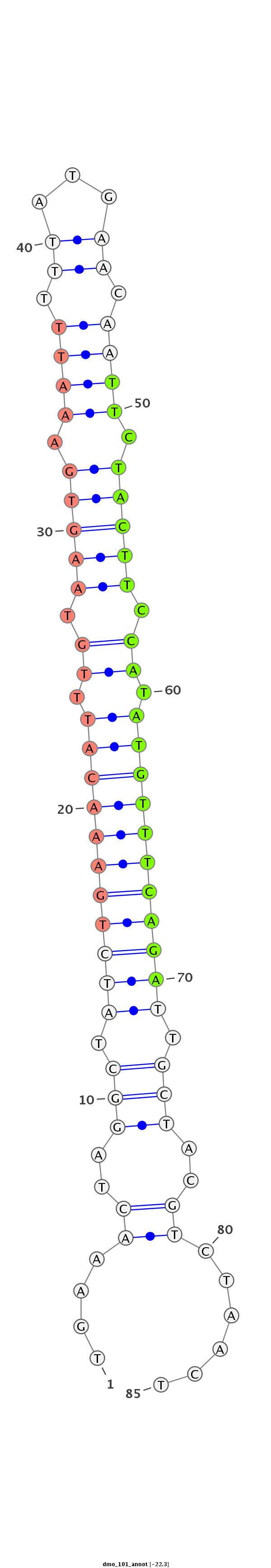
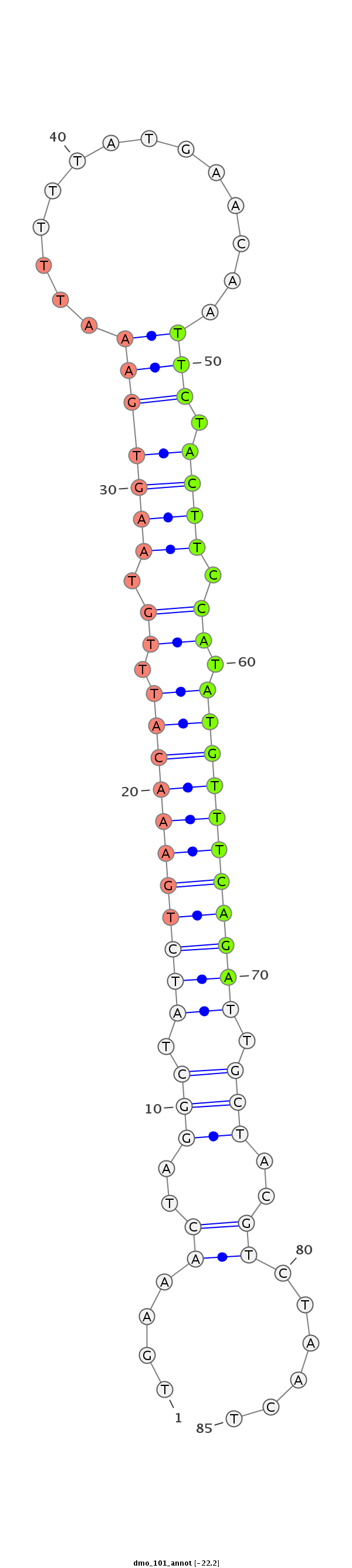
| -22.6 | -22.3 | -22.2 |
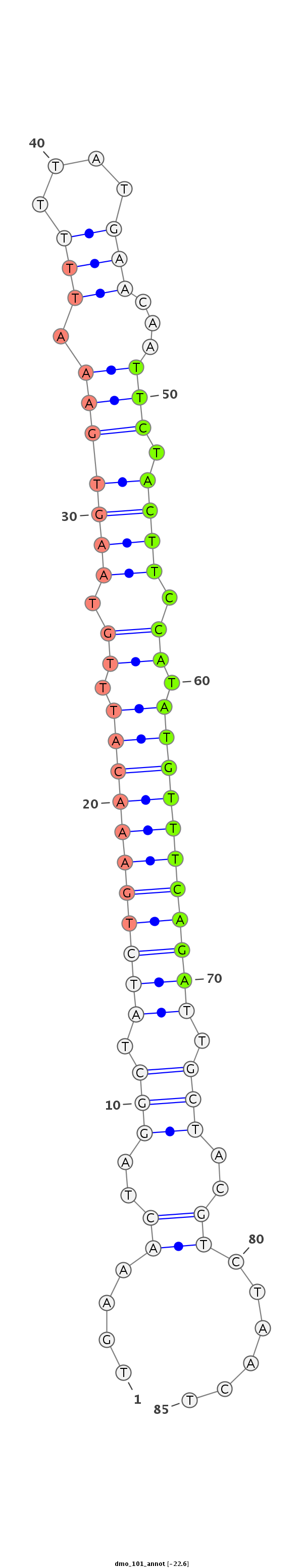 |
 |
 |
intergenic
No Repeatable elements found
|
TTCCAGATAGCTCAAATGGCAAAAGAACAACTATTTGAAACTAGGCTATCTGAAACATTTGTAAGTGAAATTTTTATGAACAATTCTACTTCCATATGTTTCAGATTGCTACGTCTAACTACTACAATCCCGATGTTCCTGCCAACTGTTTGAGCA
***********************************....((..(((.(((((((((((.((.(((((.((((.........)))).))))).)).))))))))))).)))..))......************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | V056 head |
M046 female body |
M060 embryo |
V049 head |
V041 embryo |
V110 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................TTCTACTTCCATATGTTTCAGA................................................... | 22 | 0 | 1 | 64.00 | 64 | 14 | 15 | 17 | 11 | 6 | 1 |
| ......................................................................................TACTTCCATATGTTTCAGATTG................................................ | 22 | 0 | 1 | 28.00 | 28 | 6 | 12 | 3 | 5 | 2 | 0 |
| ..................................................TGAAACATTTGTAAGTGAAATT.................................................................................... | 22 | 0 | 1 | 18.00 | 18 | 10 | 4 | 2 | 1 | 0 | 1 |
| .....................................................................................CTACTTCCATATGTTTCAGATTG................................................ | 23 | 0 | 1 | 8.00 | 8 | 1 | 3 | 2 | 0 | 2 | 0 |
| ................................................TCTGAAACATTTGTAAGTGA........................................................................................ | 20 | 0 | 1 | 6.00 | 6 | 4 | 1 | 0 | 1 | 0 | 0 |
| ...............................................ATCTGAAACATTTGTAAGTGAA....................................................................................... | 22 | 0 | 1 | 6.00 | 6 | 1 | 1 | 3 | 1 | 0 | 0 |
| ................................................TCTGAAACATTTGTAAGTGAAA...................................................................................... | 22 | 0 | 1 | 5.00 | 5 | 0 | 0 | 3 | 2 | 0 | 0 |
| ................................................TCTGAAACATTTGTAAGTGAA....................................................................................... | 21 | 0 | 1 | 5.00 | 5 | 1 | 1 | 0 | 3 | 0 | 0 |
| ...................................................................................TGCTACTTCCATATGTTTCAGA................................................... | 22 | 1 | 1 | 4.00 | 4 | 0 | 0 | 4 | 0 | 0 | 0 |
| ...................................................................................TTCTACTTCCATATGTTTCAG.................................................... | 21 | 0 | 1 | 4.00 | 4 | 2 | 0 | 0 | 2 | 0 | 0 |
| ..................................................TGAAACATTTGTAAGTGAAA...................................................................................... | 20 | 0 | 1 | 4.00 | 4 | 2 | 0 | 1 | 1 | 0 | 0 |
| ......................................................................................TACTTCCATATGTTTCAGATTGT............................................... | 23 | 1 | 1 | 4.00 | 4 | 2 | 0 | 0 | 1 | 1 | 0 |
| ......................................................................................TACTTCCATATGTTTCAGATT................................................. | 21 | 0 | 1 | 3.00 | 3 | 1 | 1 | 1 | 0 | 0 | 0 |
| ...............................................ATCTGAAACATTTGTAAGTGA........................................................................................ | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................TTATACTTCCATATGTTTCAGA................................................... | 22 | 1 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................TCTACTTCCATATGTTTCAG.................................................... | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TTAAACATTTGTAAGTGAAATT.................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................CTGAAACATTTGTAAGTGAA....................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TGAAGCATTTGTAAGTGAAATT.................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................CATTCTACTTCCATATGTTTCAGA................................................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................TTCTACTTCCATATGCTTCTG.................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................TTCTACTTCCATCTGTTTCAGA................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TGAAACATTTGTAAGTGAAAT..................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................CTACTTCCATATGTTTCAGAC.................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TTAAACATTTGTAAGTGAAAT..................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................TTCTACTTCCATATGCTTCAGA................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................ATCCGAAACATTTGTAAGTGA........................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................TCTGAAACATTTCTAAGTGA........................................................................................ | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................TCTGAAACATTTGTAAGGGAA....................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................TCTGAAACATTTGTAAGTGAAT...................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................CTACTTCCATATGTTTCCGATTG................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................TACTTCCATATGTTTAAGAATG................................................ | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................TTCTACTTCCATATGTTCCAC.................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................ATCTGAAACAGTTGTAAGTGA........................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................CTTCTACTTCCATATGTTTCAGA................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................TTCTACTTCCATATGTTTCA..................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................TATTCTACTTCCATATGTTTCAG.................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................ACTTCCATATGTTTCAGATTG................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................TTCTACTTCCATATGTTTCAGAA.................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................CTACTTCCATATGTTTCAGATTT................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................TAATTCCATAAGTTTCAGATTGC............................................... | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................TCTACTTCCATATGTTTCAGA................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TGCAACATTTGTAAGTGAAA...................................................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................TAGTTCCATATGTTTCGGCTTG................................................ | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................TGAAACATTTGTATCTGAAC...................................................................................... | 20 | 3 | 20 | 0.25 | 5 | 0 | 4 | 0 | 0 | 0 | 1 |
| ..................................................TGCCGCATTTGTAAGTGAAATT.................................................................................... | 22 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................GAAAGCCACGTCTAACTAC.................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
|
AAGGTCTATCGAGTTTACCGTTTTCTTGTTGATAAACTTTGATCCGATAGACTTTGTAAACATTCACTTTAAAAATACTTGTTAAGATGAAGGTATACAAAGTCTAACGATGCAGATTGATGATGTTAGGGCTACAAGGACGGTTGACAAACTCGT
************************************....((..(((.(((((((((((.((.(((((.((((.........)))).))))).)).))))))))))).)))..))......*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V049 head |
M046 female body |
V110 male body |
|---|---|---|---|---|---|---|---|---|
| ..................................................ACTTTGTAAACATAGACTTG...................................................................................... | 20 | 3 | 20 | 0.15 | 3 | 0 | 2 | 1 |
| .....................................................TTGTAAAAATTCACTTTCATAAT................................................................................ | 23 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 |
| ......................................................TGTAAAAATTCACTATCAAAAT................................................................................ | 22 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droMoj3 | scaffold_6328:351285-351440 + | dmo_101 | confident | TTCCAGATAGCTCAAATGGCAAAAGAACAACTATTTGAAACTAGGCTATCTGAAACATTTGTAAGTGAAATTTTTATGAACAATTCTACTTCCATATGTTTCAGATTGCTACGTCTAACTACTACAATCCCGATGTTCCTGCCAACTGTTTGAGCA |
| droGri2 | scaffold_15110:14614387-14614434 + | -------------------------AACAATTTTTTGGGTTTAGACTAATAAAAATGTTTTTGAGCGAAATTT----------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 10/20/2015 at 06:47 PM