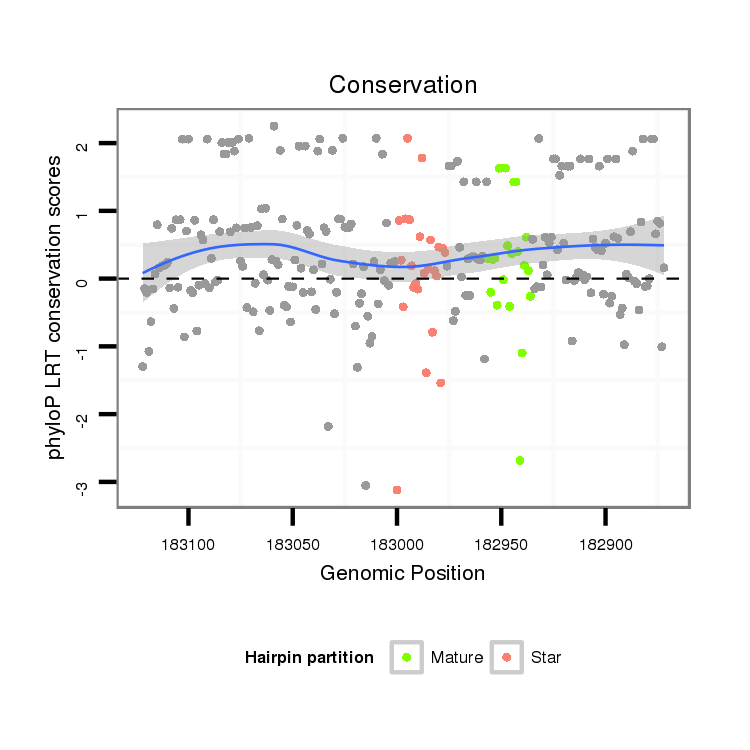
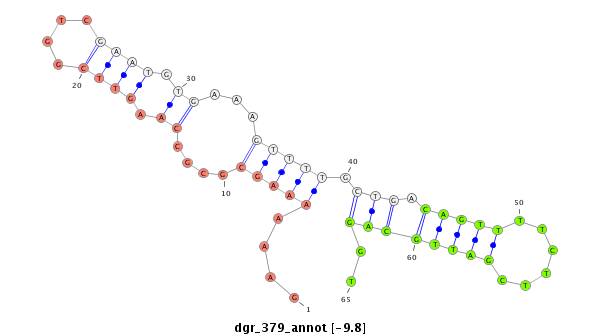
| droGri2 |
scaffold_15126:182872-183122 - |
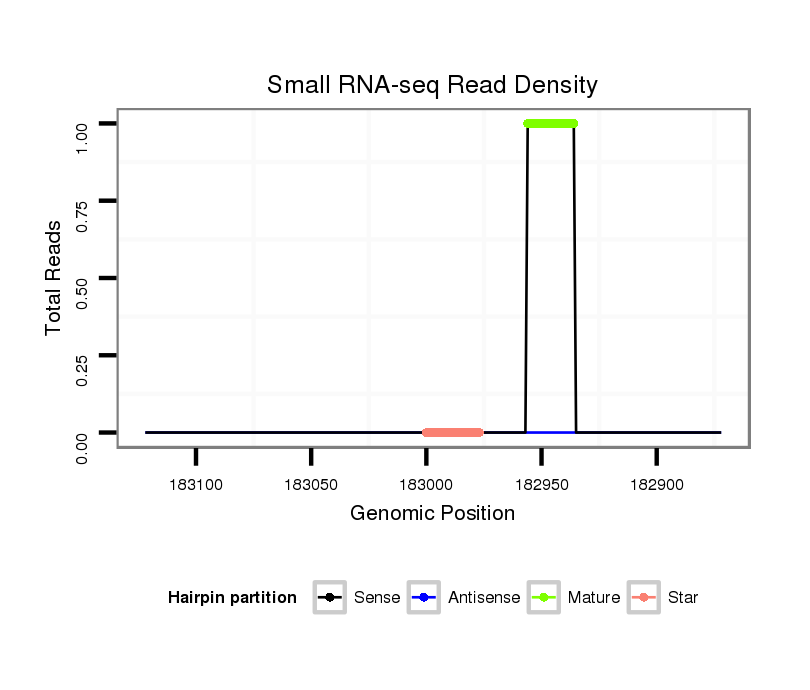
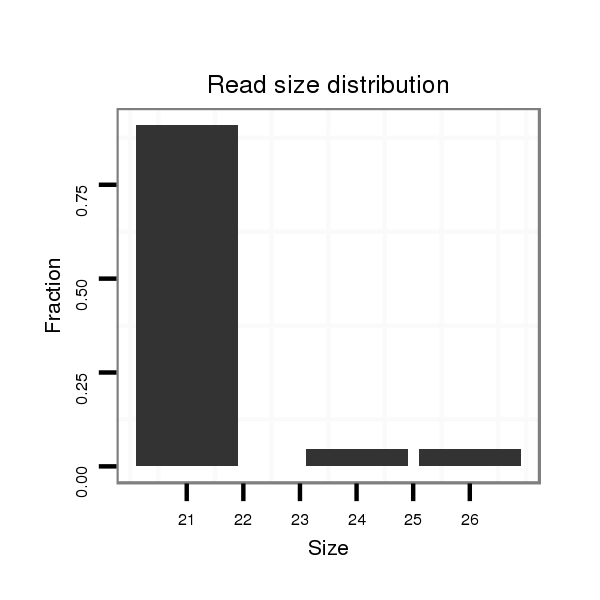
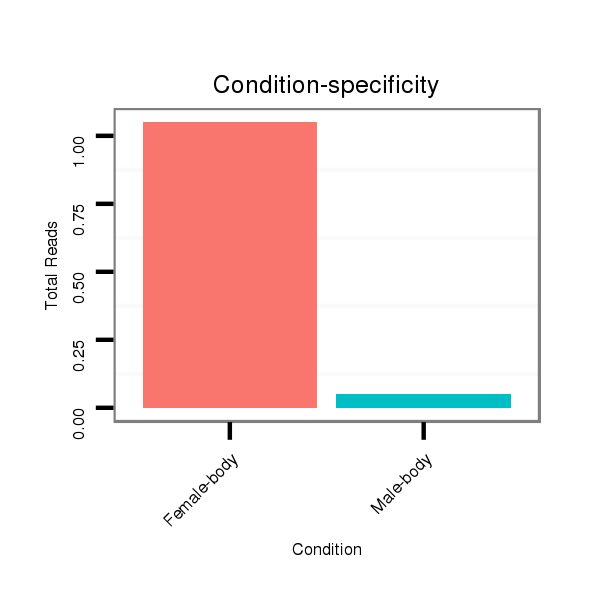
dgr_379 |
GTCCG-CG-ACGACCC-AAATTTGCTCCAGAGGGTCATAACTGGTGACGAATCGTGAGTTTATGGTTACGACGTGGAAACCAAAGCTCAATCATCTCAATGGAAGCTGC-CGCACGAA-----CCAAGACCGAAAAAAGCG--------------------------------CGCCAAGTTCGGTCG-AATG---TGAAAGTTTTGCTGACAGTTTTCTTCGATTG------------------------------------------CAGGTGCTTCG-------AAGATTGGAAAAACCGTTGGCACAAGTGTAT--------AATATCTCATGGGGATTA------CTTTGA-----------------------------------------AGG |
| droMoj3 |
scaffold_6540:8802936-8803030 + |
|
AAAGT-T-----------------------------------------------------TTTTTTT-----------------------------------------------------------------------------------------------------------------------------------------------------TTTTTTT----------------------------------------TTTGCCTAGGCTTTTCG-------AAGATTGGAAGAAATATTGTTAAAGGTAGAT--------ATTATTTGATAGGGTTTG------CTTTGA-----------------------------------------ATG |
| droWil2 |
scf2_1100000004595:101888-102078 + |
|
TTCGC----GTGGTCC-AGACTTCTTGAATCGCATTATTACTGGTGACGAGACATGGGTCTATGAGTTCGACACCTTAACAAGCCAGCAATCGTCTGAATGGCGAATGG-AAAACGAG-----ACAAATCCCAAAAAAGCG--------------------------------CACCGAATCCGCTCG-AAAT---TGAAGGCTTTGCTGACTGTGTTCTTCGATTT------------------------------------------CTGGGGTGTCG------------------------------------------------------------------------------------------------------------------------ |
| droAna3 |
scaffold_13034:1201891-1202164 - |
|
ATCAA-CG-ACGACCC-AGATTTACTCAAAAGGGTCATAACTGGTGACGAATCATGGGTATATGGTTATGATATCGAAACCAAAGCCCAATCGTCCCAATGGAAGAGCAACTGAAGAG-----A-----CCTATGAAAGGACGGAGATTTGCAACGATTAAAGAGATAAACCCTGC---------------------------------------------------ATCGCTGCAAGAGCTCAAGGCTATACCGAAAAGTGCTTATGGGAAGTGCTTTG-------AGGATTGGAAAAACCGTTAGCATAAGTGTAT--------TGTATTTGAGGGGGATTA------CTTTGA-----------------------------------------AAG |
| droBip1 |
scf7180000396324:44149-44394 - |
|
GTCAA-CG-ACGGCCG-AGATTTACTCAAAAGGGTCATAACTGGTGACGAATCATGTGTATATGATTATGATATCGAAAACAAAGCCCAATCGTCCCAATGGAAGAGCC-CAGGTGAG-----CCAAGACCGAAAAAAGCA--------------------------------CGCATGGCTG---------------------------------------------------GAAGATCTCAAGGCTATACCGAAAAGTTCTTATGGGAAGTGCTTTT-------AGGATTGGAAAAACCGTTGGAATTATTGTAT--------TGTATCTGAGGGGGATTA------CTTTGA-----------------------------------------AGG |
| droKik1 |
scf7180000302223:15869-15933 + |
|
TAT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCG-------AGGATTTGAAAAAACGCTGGCATAAGTGTAT--------GTTATCGGGAGGGGATTA------CTTTCA-----------------------------------------AGG |
| droFic1 |
scf7180000449169:6634-6920 - |
|
GTCCG-CG-ACGACCCAAAATTTGCTCCAGAGGGTTATAGCTGGTGACGAATCGTGGGTTTATGGTTACGACGTGGA-------GCTCATTCATCTTAATGGAAGCTGC-CGCACGAA-----CCAAGACCGAAAAAAGCG--------------------------------CGCCAAGTTCGGTCG-AATG---TGAAAGTTTTGCTGACAGTTTTCTTCGATTCATCGAAGGAGAAGCTGAACAAGATAACAAAAAATGATTTTTTGAAGTGCTTCG-------AAGATTGGAAAAACAGTTGGCACAAGTGCAT--------AGTATCTCATGGGGATTA------ATTTGA-----------------------------------------AGG |
| droEle1 |
scf7180000489729:28143-28367 + |
|
GTCAC-TT-ACGAACC-AAACTTACTCAAAAGGGTCATAGCTGGTGACGAATCATGGGTACATGGTTGTGATATCGAAACCAAAGCCCAATCCTCCCAATGGAAGAGCC-CAGGTGAG-----CCAAGAACACGCAAAACA----------------------------------------------------ACTTGT----------------------------AACCTTCACGTCTCTCACAACCATACAAAAAA-----------------------------------------------------------------------------GGGGATTC------ACTTGAAACTTGGTACAGATGTTAGTGAAGAGTGTACCAACATAAT---A |
| droRho1 |
scf7180000761329:22138-22388 + |
|
GTCAA-CG-ACGACCC-AGATTTACTCAAAAGGGTCATAACTGGTGACGAATCATGGGTATATGGTTATGATATCGAAACCAAAGCCCAATCGTCCCAATGGAAGAGCC-CAGGCGAG-----CCAAGACCGAAAAAAGCA------------------------AAAAAT--CGCCGAGCTCGC--AAAAGAACTTGT----------------------------AAC-TTTACGCCTCTCACAACCAAACAAAAGA-----------------------------------------------------------------------------GGATGTTC------CATTGAAACTTGGTACAGATGTTAGGGAAGAGTGTACCAATATAACAAAA |
| droBia1 |
scf7180000297034:27704-27960 - |
|
GTCAA-CG-ACGACTC-AGATTTACTCAAAAGGGTCATAACTGGTGACGAATCATGGGTATATGGTTATGATATCGAAACCAAAGCCCAATCGTCACAATGAAAGAGCC-CAGGTGAG-----CCAAGACCGAAAAAAGCA--------------------------------CGCCAAGTTCGATCA-AATG---TCAAAGTTTTGATCACTGTATTCTTCGATTA------------------------------------------CCATGGCGTGGTGCATCAGGAGTT---------CTTACCATA------TGGTATTCCAGTATTATGTGGAAGTTATGCGCCGTTTGC-----------------------------------------GAG |
| droEug1 |
scf7180000405005:1283-1534 + |
|
GTCAA-CG-ACGACCC-AGATTTACTCAAAAGGGTCATAACTGGTGACGAATCATGGGTATATGGTTATGATATCGAAACCAAAGCCCAATCGTCCCAATGGAAGAGCC-CAGACGAG-----CCAAGACCGAAAAAAGCA------------------------AAAAAT--CGCCGAGCTCGATCA-AATAACTTGT----------------------------AAC-TTTACGCCTCTCACAACCAAACAAAAGA-----------------------------------------------------------------------------GGATGTTC------CATTGAAACTTGGTACAGATGTTAGGGAAGAGTGTACCAATATAACAAAC |
| droSim2 |
node_97685:646-727 + |
|
------------------------------------------------------TGGGTTTATGGTTATGAAGTGGAAACCAAAGCTCAATCATCTCAATGGAAGCTGC-CGCACAAAGGTGCCCTAAACTGAAGAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droSec2 |
scaffold_295:15064-15290 + |
|
ATTCAACA-ACCACCG-AATTCTCCTGACTTGGCTCCCTG-------CGACTTTTTCCTATTC------GGTCGACTCAAGAAAC-------CGCTCCGTGGAACACGT-TTCAGCAC-----CCGGGATGAAATCATGGA------------------------A-----------------------------------------------------------AAATCGAAGATGGCTTTGATGGCCATACCGCAAATTGAATATGAAAAATGTTTCA-------AGGATTGGATCAAGCGCTGGCATAAGTGCGT--------TGCAGTCGATGGGGAGTA------CTTTGA-----------------------------------------AGG |
| droYak3 |
3R:815707-815898 - |
|
CAACA-A--AAGACCC-GAACTTTCTTTACAAGATTGTGAATGGTGACGAAACGTGCTATTTTAAATATGATCCCCAAACGAAACGTCAGAGTGCTGAATAGAAGGTGC-CCGGCGAG-----CCGAAACCTAAAAAATTA--------------------------------AGCATGGAGAAGCTA-AAAG---TGAAGACAATGCAGATTTGTTTTTATGATTC------------------------------------------CAAGTGTATTG------------------------------------------------------------------------------------------------------------------------ |
| droEre2 |
scaffold_4929:21529099-21529283 + |
|
GCAAA-AATAATACAT-GAACTTTCTTTACGAGATTATGACTGGTAACAAAACGTGGAATTTTCAATATGACCCCGAAACGAAACGTCAGAGTACTGAATGGAAGGCGC-CGGACGAG-----CCAAAGCCCAAAAAATCG--------------------------------GGCCTGGAGAAGTCA-AAAG---TGAAGGCAATACTTATTTGTTTTTAAGATTT------------------------------------------CA--------------------------------------------------------------------------------------------------------------------------------- |