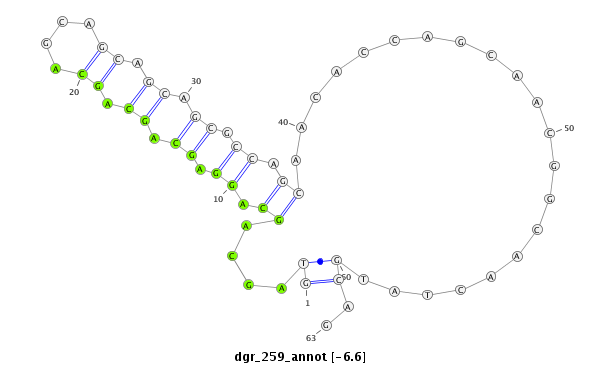
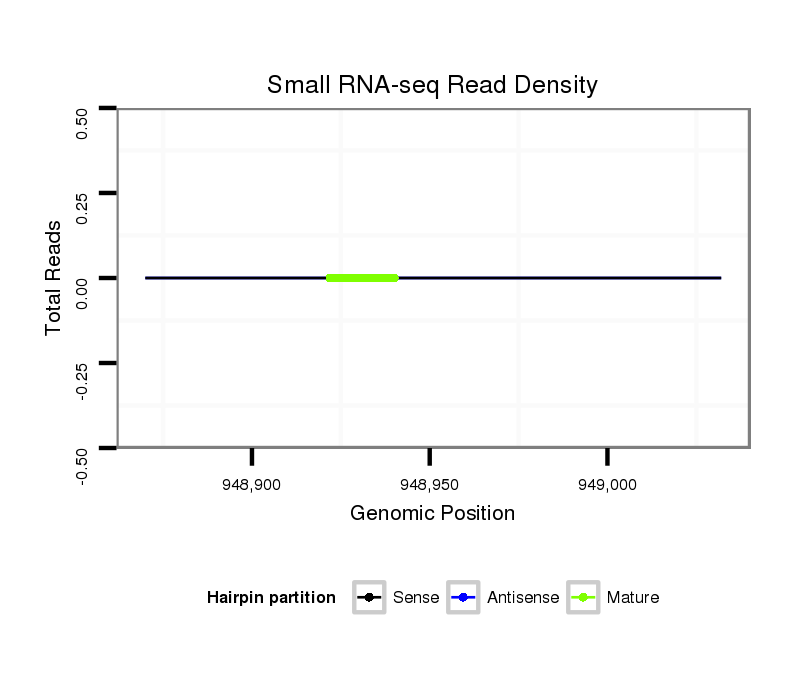
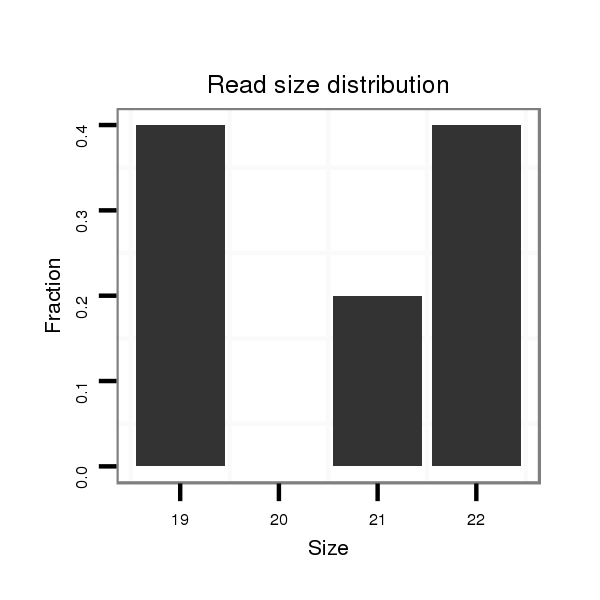
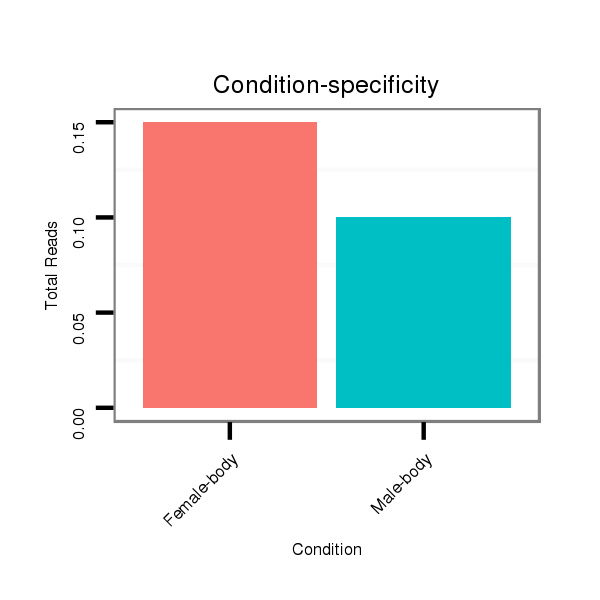
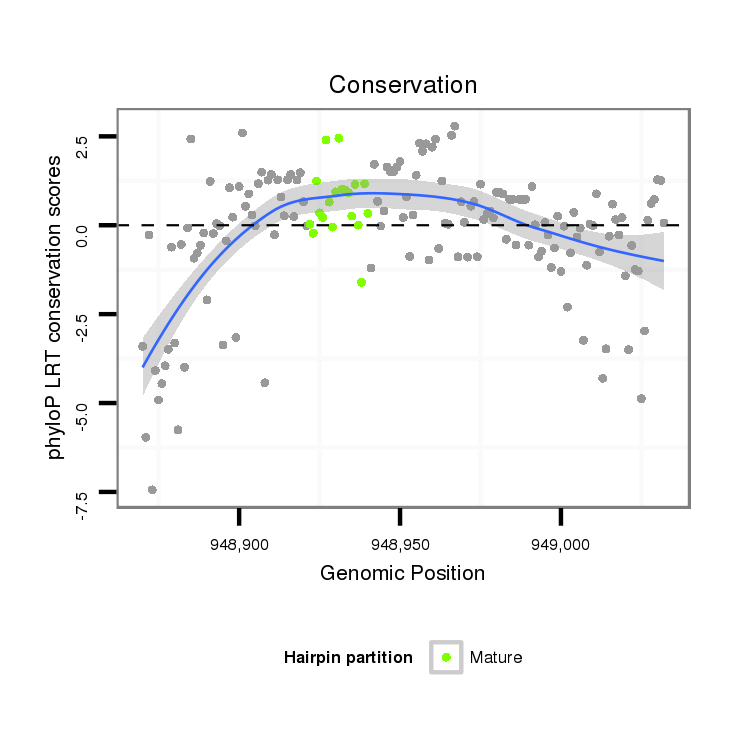
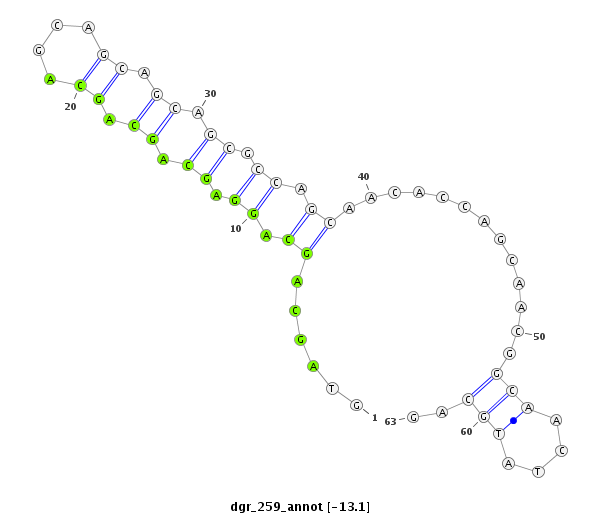
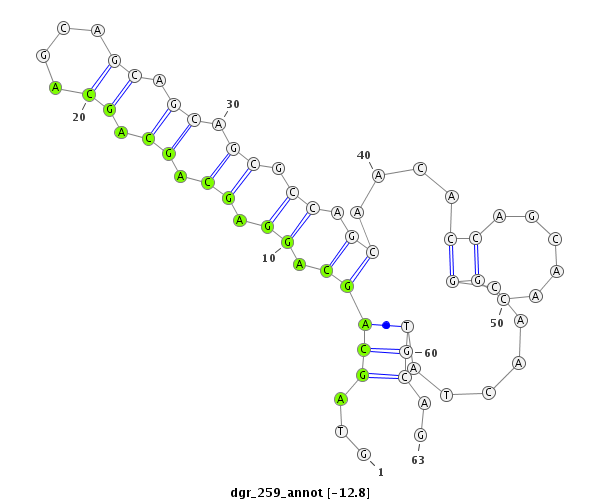
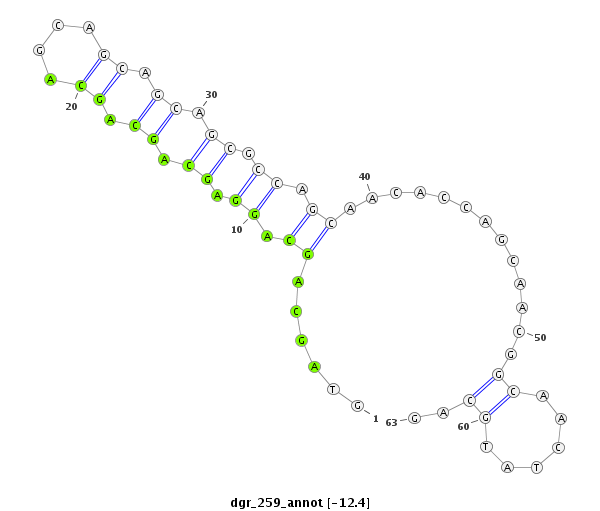
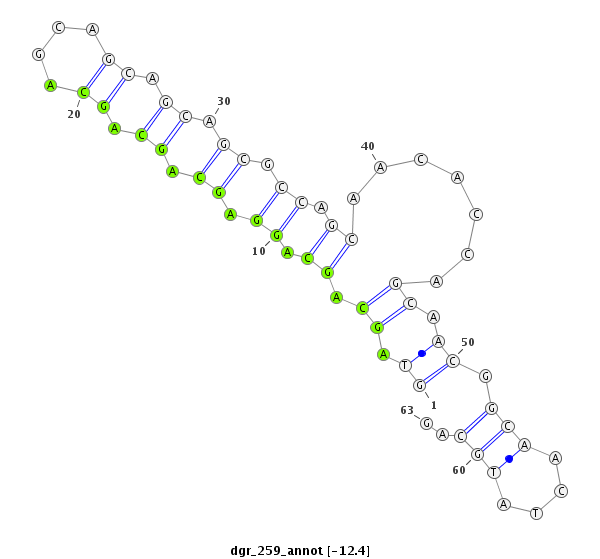
| CTCGCCCAAGAGCAGCAGCA-GCAG------------CAGCAG--CATCA---G---CAG---CAAC---------AGCAGCA------GCA---------------GCAG------------------CAGCAG---------CCACAGCA------GC------AGCAGCAGC------ATCAGCAGCATCAGCA---------------------------------------------------GCAGCGG-------------------------------------CAGCAGC---------GGCAGCA-----------GCGACC--AG------------CCCACAC--ACAGCAACATTCCGCGCAGCA | Size | Hit Count | Total Norm | Total | M055
Female-body | M057
Embryo | V060
Head | V040
Embryo | V107
Male-body | V108
Head | GSM1528801
follicle cells |
| ..CGCCCAAGAGCAGCAGCA-GC................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................GCA-GCAG------------CAGCAG--CATCA---G---CAG---CA......................................................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.55 | 11 | 8 | 0 | 0 | 0 | 2 | 1 | 0 |
| .....................................................................................................................................................GCA------GC------AGCAGCAGC------ATCAGCAGCA........................................................................................................................................................................................... | 24 | 20 | 0.55 | 11 | 8 | 0 | 0 | 0 | 2 | 1 | 0 |
| .........GAGCAGCAGCA-GCAG------------CAGCAG--CATCA---G---CA............................................................................................................................................................................................................................................................................................................................... | 29 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................A------GC------AGCAGCAGC------ATCAGCA.............................................................................................................................................................................................. | 19 | 20 | 0.45 | 9 | 0 | 0 | 0 | 0 | 2 | 7 | 0 |
| .......................AG------------CAGCAG--CATCA---G---CAG---CA......................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.40 | 8 | 0 | 0 | 0 | 0 | 3 | 5 | 0 |
| ......................................................................................................................................................................AGCAGCAGC------ATCAGCAGCA........................................................................................................................................................................................... | 19 | 20 | 0.40 | 8 | 0 | 0 | 0 | 0 | 3 | 5 | 0 |
| ...................A-GCAG------------CAGCAG--CATCA---G---CA............................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................CCACAGCA------GC------AGCAGCAGC------AT................................................................................................................................................................................................... | 21 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................G------------CAGCAG--CATCA---G---CAG---CAAC---------AGCAGCA------................................................................................................................................................................................................................................................................................................. | 27 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............GCAGCA-GCAG------------CAGCAG--CATCA---G---CA............................................................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............AGCAGCA-GCAG------------CAGCAG--CATCA---G---CAG---CA......................................................................................................................................................................................................................................................................................................................... | 28 | 18 | 0.22 | 4 | 2 | 0 | 0 | 0 | 1 | 1 | 0 |
| ................................................................AAC---------AGCAGCA------GCA---------------GCAG------------------C........................................................................................................................................................................................................................................................ | 18 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 0 | 4 | 0 |
| .............................................CATCA---G---CAG---CAAC---------AGCAGCA------GCA---------------GCAG------------------......................................................................................................................................................................................................................................................... | 27 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............GCAGCA-GCAG------------CAGCAG--CATCA---G---CAG---CA......................................................................................................................................................................................................................................................................................................................... | 27 | 19 | 0.16 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................AGCA-GCAG------------CAGCAG--CATCA---G---CAG---CA......................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.15 | 3 | 1 | 0 | 0 | 0 | 0 | 2 | 0 |
| .....................GCAG------------CAGCAG--CATCA---G---CA............................................................................................................................................................................................................................................................................................................................... | 18 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 3 | 0 |
| ....................................................................................................................................................AGCA------GC------AGCAGCAGC------ATCAGCA.............................................................................................................................................................................................. | 22 | 20 | 0.15 | 3 | 1 | 0 | 0 | 0 | 0 | 2 | 0 |
| .................GCA-GCAG------------CAGCAG--CATCA---G---C................................................................................................................................................................................................................................................................................................................................ | 20 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................AGCA------GC------AGCAGCAGC------ATCAGCAGCA........................................................................................................................................................................................... | 25 | 20 | 0.15 | 3 | 1 | 0 | 0 | 0 | 0 | 2 | 0 |
| ............................................................................................................................................................................AGC------ATCAGCAGCATCAGCA---------------------------------------------------GCAG.............................................................................................................................. | 23 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............AGCAGCA-GCAG------------CAGCAG--CATCA---G---CA............................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................GC------AGCAGCAGC------ATCAGCAGCA........................................................................................................................................................................................... | 21 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| .......................................................................................................................................................A------GC------AGCAGCAGC------ATCAGCAGCA........................................................................................................................................................................................... | 22 | 20 | 0.10 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............AGCAGCA-GCAG------------CAGCAG--CATC......................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| .....................................................................................................................................................GCA------GC------AGCAGCAGC------ATCAGCA.............................................................................................................................................................................................. | 21 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| .................GCA-GCAG------------CAGCAG--CATCA---G---CAG---C.......................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............CAGCA-GCAG------------CAGCAG--CATCA---G---CA............................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| .....................................................................................................................................................GCA------GC------AGCAGCAGC------ATCAGCAGC............................................................................................................................................................................................ | 23 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................AGCAGCATCAGCA---------------------------------------------------GCAGC............................................................................................................................. | 18 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................AGCAG---------CCACAGCA------GC------AGCA................................................................................................................................................................................................................ | 19 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................AGCAGC------ATCAGCAGCATCAGCA---------------------------------------------------GC................................................................................................................................ | 24 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................AGCAGC------ATCAGCAGCATCAGC...................................................................................................................................................................................... | 21 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............GCAGCA-GCAG------------CAGCAG--CATCA---G---................................................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................CAGCA---------------------------------------------------GCAGCGG-------------------------------------CAGCAGC---------...................................................................... | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................CAAC---------AGCAGCA------GCA---------------GCAG------------------CAGCA.................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................G------------CAGCAG--CATCA---G---CAG---CAA........................................................................................................................................................................................................................................................................................................................ | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................AGCA-GCAG------------CAGCAG--CATCA---G---C................................................................................................................................................................................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................C------AGCAGCAGC------ATCAGCAGCA........................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................AGCA------GC------AGCAGCAGC------ATCAGC............................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................C------ATCAGCAGCATCAGCA---------------------------------------------------GC................................................................................................................................ | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............CAGCAGCA-GCAG------------CAGCAG--CATCA---G---CA............................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................CAG------------CAGCAG--CATCA---G---CAG---CA......................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................GCA-GCAG------------CAGCAG--CATCA---G---CA............................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................AGCA------GC------AGCAGCAGC------ATCAG................................................................................................................................................................................................ | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................AGCA-GCAG------------CAGCAG--CATCA---G---CA............................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................GCA------GC------AGCAGCAGC------ATCAGC............................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................CA-GCAG------------CAGCAG--CATCA---G---CAG---CA......................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................GC------AGCAGCAGC------ATCAGCAGC............................................................................................................................................................................................ | 20 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................G---CAAC---------AGCAGCA------GCA---------------GCA............................................................................................................................................................................................................................................................................ | 18 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................CA-GCAG------------CAGCAG--CATCA---G---CAG---........................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............GCAGCA-GCAG------------CAGCAG--CATC......................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................A-GCAG------------CAGCAG--CATCA---G---CAG---CA......................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............GCAGCA-GCAG------------CAGCAG--CATCA---G---CAG---C.......................................................................................................................................................................................................................................................................................................................... | 26 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............AGCAGCA-GCAG------------CAGCAG--CATCA---G---C................................................................................................................................................................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........GCAGCAGCA-GCAG------------CAGCAG--CATCA---..................................................................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |